Method for identifying structure of yeast colony of Daqu starter or fermented grain of distilled spirit by using denaturing gradient electrophoresis
A technique of denaturing gradient and electrophoresis, which is applied in the direction of microorganism-based methods, biochemical equipment and methods, and microorganism determination/inspection, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
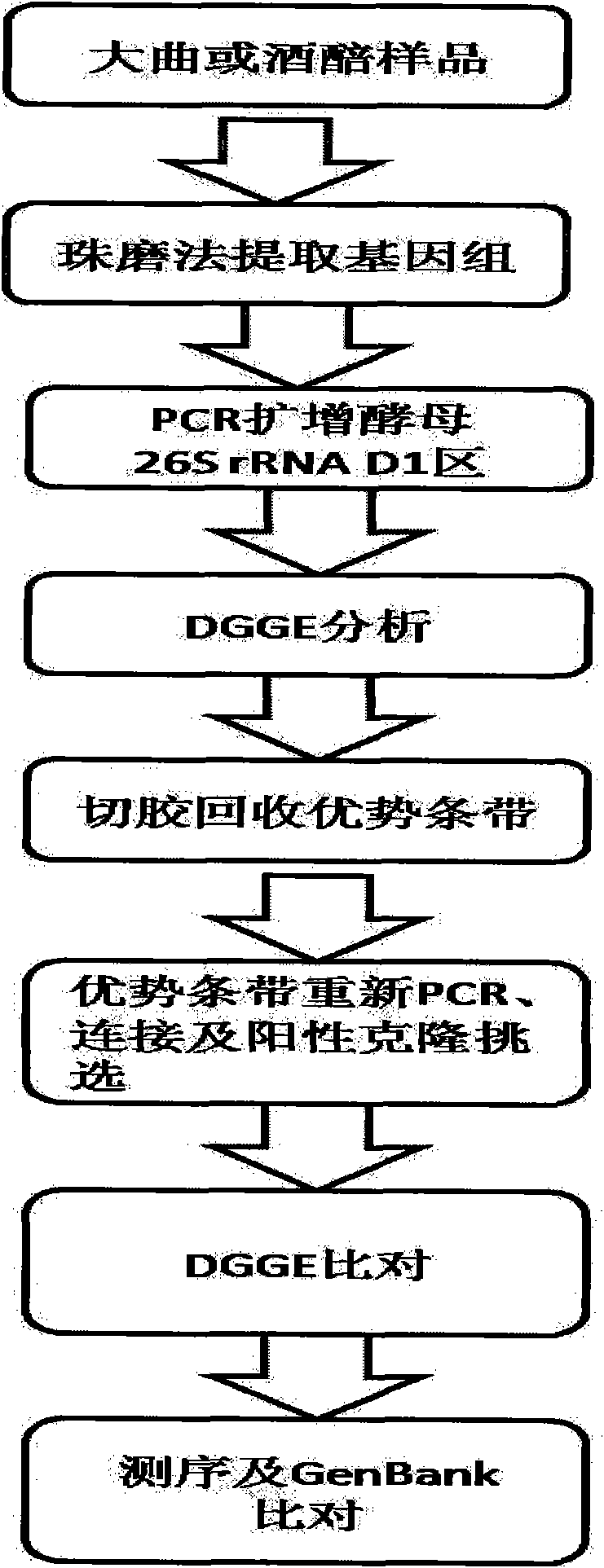
[0027] Daqu, a strong-flavor liquor from northern Jiangsu, was used to sample the center and skin of the koji. The genomes of all microorganisms in the samples were extracted by the Beadbeater method. Use a Bio-rad PCR instrument for PCR amplification. The yeast PCR conditions are as follows: use universal primers to amplify the DNA fragments in the yeast 26S rDNAD1 region. The primers are:
[0028] NL1-GC: 5'-CGCCCGCCGCGCGCGGCGGGCGGGGCGGGGGCATATCAATAAGCGGAGGAAAAG-3',
[0029] LS2: 5'-ATTCCCAAACAACTCGACTC-3';
[0030]The 25 μL reaction system includes 2.5 μL 10×Buffer, 2 μL 25 mmol / L dNTP mixture, 1U TaqDNA polymerase, 25 pmol of each primer, about 10 ng of DNA template, and 25 μL of double distilled water. PCR reaction program: 94°C pre-denaturation for 4 min; 94°C for 1 min, 65°C for 1 min, 72°C for 1 min, 30 cycles; finally 72°C for 10 min.
[0031] The DGGE conditions are: the acrylamide gel concentration is 8%, the denaturing gradient range is 10%-50% (aqueous solution...
Embodiment 2
[0034] Fen-flavor liquor fermented grains were used, and samples were taken at three points, the upper, middle, and lower points in the fermentation vat, and mixed after the sampling was completed. The genomes of all microorganisms in the samples were extracted by the Beadbeater method. PCR amplification was performed using a Bio-rad PCR instrument. Yeast PCR conditions are as follows: use universal primers to amplify the DNA fragment of yeast 26S rDNAD1 region. Primers are:
[0035] NL1-GC: 5'-CGCCCGCCGCGCGCGGCGGGCGGGGCGGGGGCATATCAATAAGCGGAGGAAAAG-3',
[0036] LS2: 5'-ATTCCCAAACAACTCGACTC-3';
[0037] The 25 μL reaction system includes 2.5 μL 10×Buffer, 2 μL 25 mmol / L dNTP mixture, 1U TaqDNA polymerase, 25 pmol of each primer, about 10 ng of DNA template, and 25 μL of double distilled water. PCR reaction program: 94°C pre-denaturation for 4 min; 94°C for 1 min, 65°C for 1 min, 72°C for 1 min, 30 cycles; finally 72°C for 10 min.
[0038] The DGGE conditions are: the acryl...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com