Chimeric primers for improved nucleic acid amplification reactions
一种扩增反应、嵌合引物的技术,应用在核酸扩增领域,能够解决寡核苷酸引物不能被完全复制等问题
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
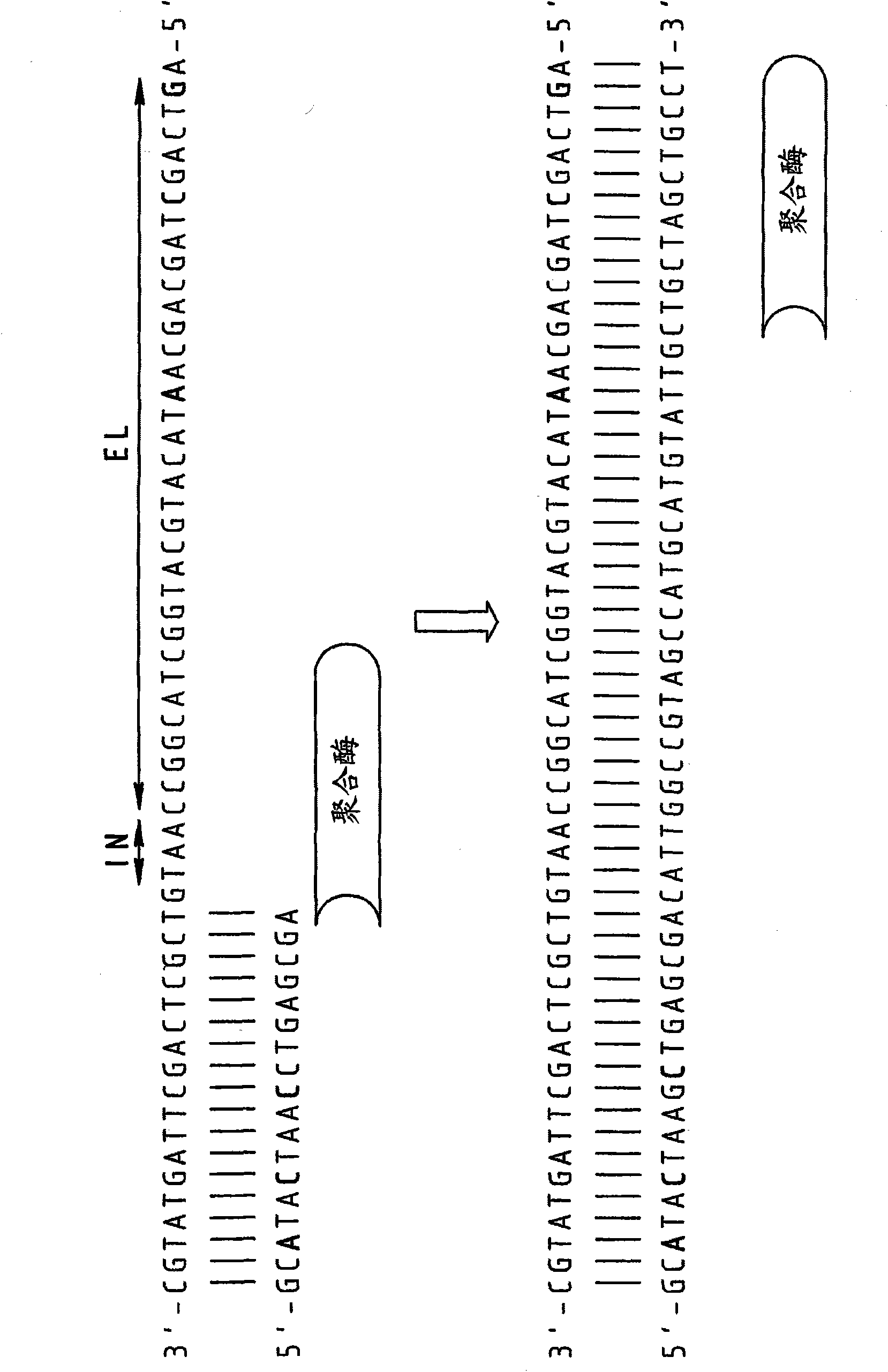
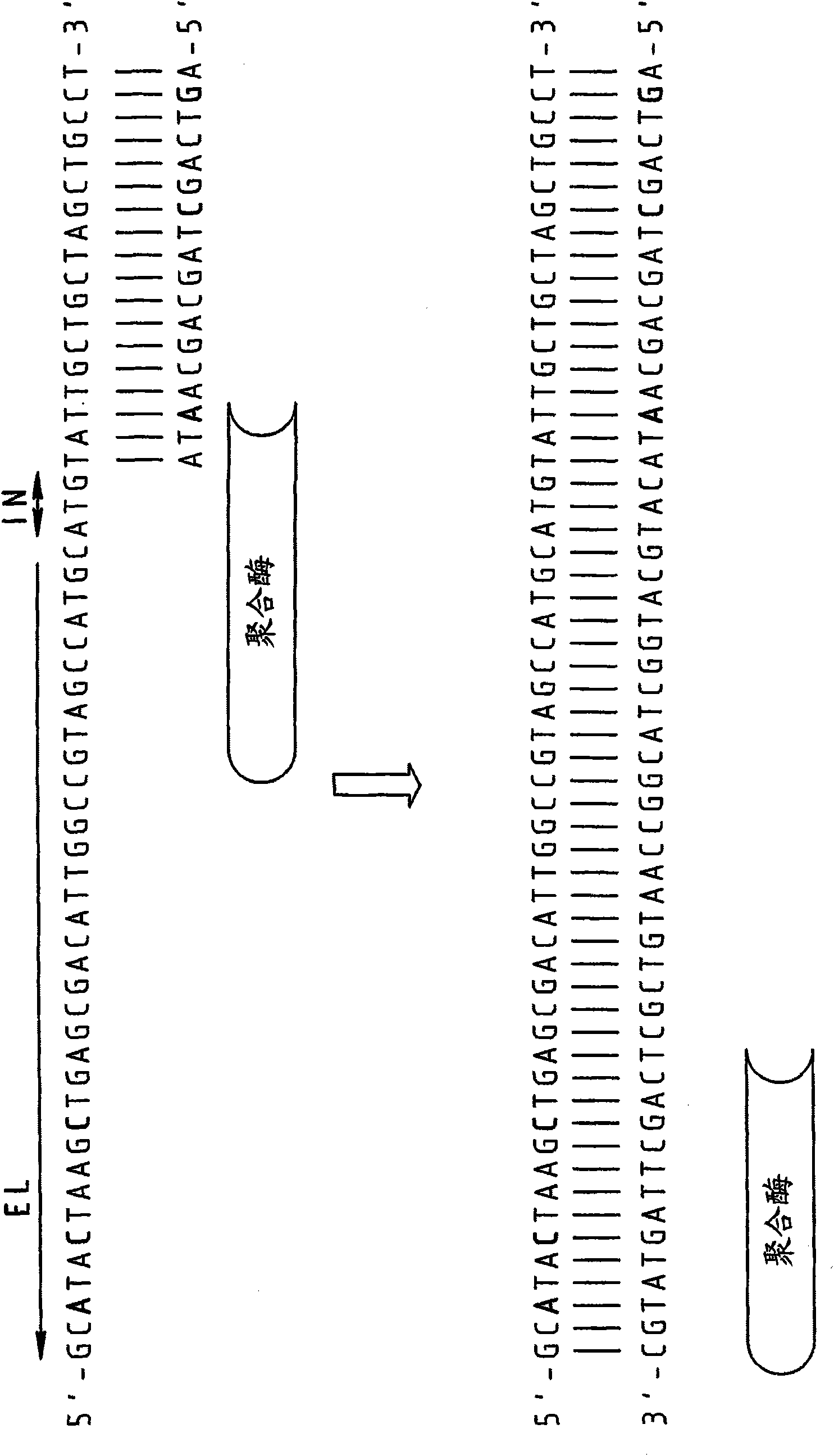
[0089] to 10 4 Copies of the 16s RNA gene of strain E. canis (EC) were subjected to real-time PCR using DNA primers and RNA / DNA chimeric primers.
[0090] DNA primers:
[0091] Forward primer: 5'-TCGCTATTAGATGAGCCTACGT3' (SEQ ID NO: 4)
[0092] Reverse primer: 5'-GAGTCTGGACCGTATCTCAGTT-3' (SEQ ID NO: 5)
[0093] RNA / DNA Chimeric Primers
[0094] (embedded RNA bases are in bold):
[0095] Forward primer: 5'-TCGCUATUAGATGAGCCUACGT-3' (SEQ ID NO: 6)
[0096] Reverse primer: 5'-GAGTCTGGACCGUATCTCAGTT-3' (SEQ ID NO: 7)
[0097] The results are presented in Figure 5 middle. As can be seen, when DNA primers were used, in the absence of template, obvious artifacts could be detected after 27 cycles (see non-templated control (NTC) curve), but when chimeric primers were used Artifacts remained below the detection threshold. In fact, the C(t) value of the specific template-dependent product was delayed by about 3 cycles, but the elimination of the non-specific product exten...
Embodiment 2
[0100] Real-time PCR was performed on the 16s RNA gene of the bacterial strain Ehrlichia canis (EC) with a series of dilutions, using the same DNA and chimeric primers as in Example 1. The results are presented in Figure 7A and 7B middle. An additional real-time PCR was performed on a series of dilutions of the hsp70 gene of the strain Babesia canis (BC) using the following primers and the results are presented in Figure 8A and 8B middle.
[0101] DNA primers:
[0102] Forward primer: 5'-GTCATCACTGTGCCTGCGTACT-3' (SEQ ID NO: 8)
[0103] Reverse primer: 5'-GCATGACGTTGAGACCGGCAAT-3' (SEQ ID NO: 9)
[0104] RNA / DNA chimeric primers:
[0105] (embedded RNA bases are in bold):
[0106] Forward primer: 5'-GTCATCACTGTGCCTGCGUACT-3' (SEQ ID NO: 10)
[0107] Reverse primer: 5'-GCATGACGTTGAGACCGGCAAT-3' (SEQ ID NO: 11)
[0108] This example shows that the sensitivity of the amplification reaction is increased when RNA / DNA chimeric primers are used. In both cases, the us...
Embodiment 3
[0110] In this example, serial dilutions of the canine ACTB gene were subjected to qPCR using DNA and chimeric primers, followed by Tm analysis.
[0111] DNA primers:
[0112] Forward primer: 5'-GCGCAAGTACTCTGTGTGGAT-3' (SEQ ID NO: 12)
[0113] Reverse primer: 5'-GTCGTACTCCTGCTTGCTGAT-3' (SEQ ID NO: 13)
[0114] RNA / DNA chimeric primers:
[0115] (embedded RNA bases are in bold):
[0116]Forward primer: 5'-GCGCAAGUACTCTGTGTGGAT-3' (SEQ ID NO: 14)
[0117] Reverse primer: 5'-GTCGUACTCCTGCTTGCTGAT-3' (SEQ ID NO: 15)
[0118] This example demonstrates the improved specificity obtained by using PCR chimeric primers prior to HRM analysis at low template concentrations. When using DNA primers ( Figure 9A ), in the Tm diagram, it can be seen that the non-specific product peak is in the range of 70°C to 78°C, while the Tm peak of the expected amplicon in the high concentration template is in the range of 81°C to 82°C. Figure 9B Tm plots obtained when using DNA / RNA chimeri...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com