Method for quickly constructing gene targeting vector based on site specificity recombination
A carrier and site technology, applied in the fields of biotechnology and genetic engineering, can solve the problems of low homologous recombination efficiency and specificity that need further research, and achieve the effect of rapid construction and simple construction method
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
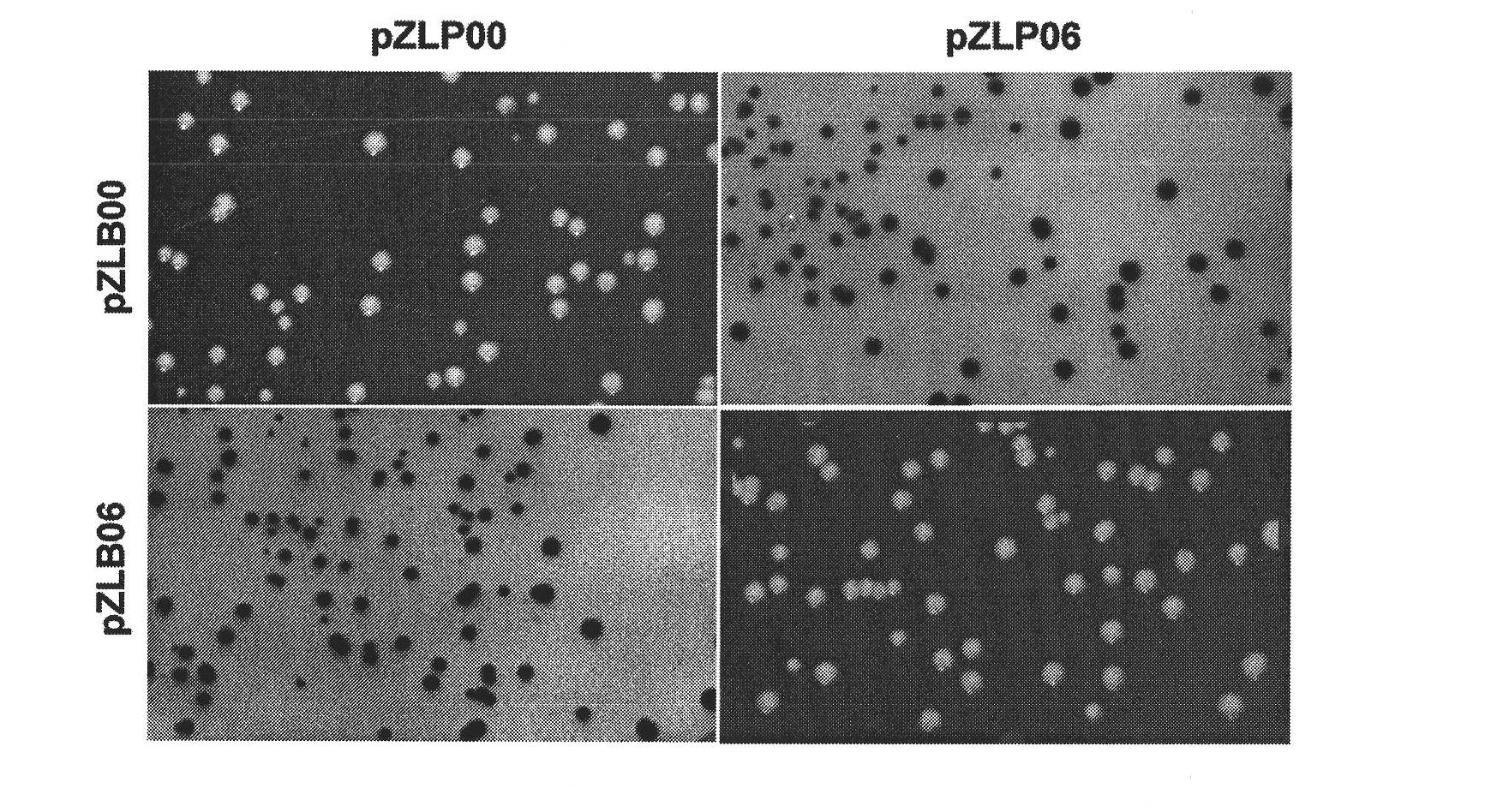
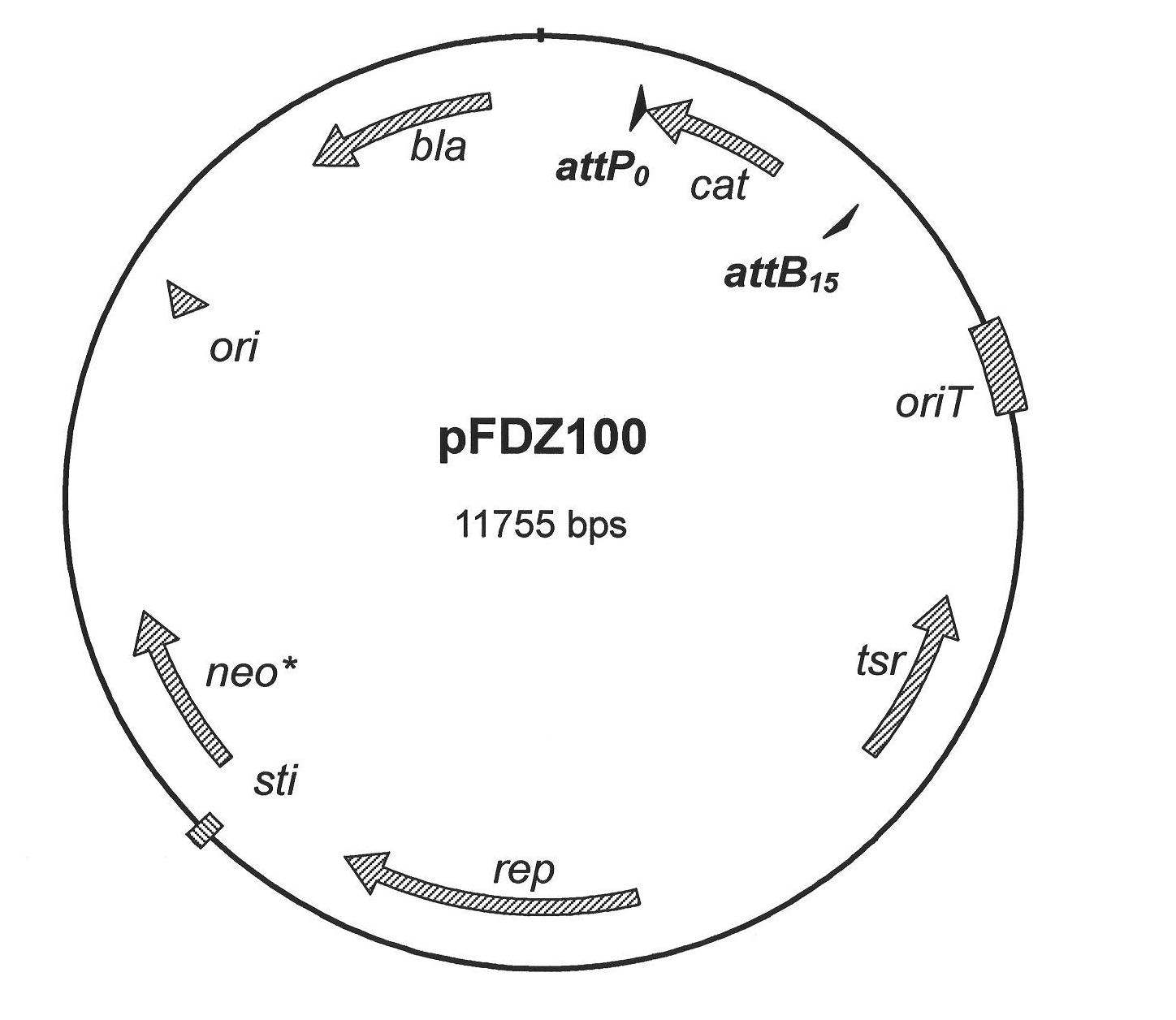
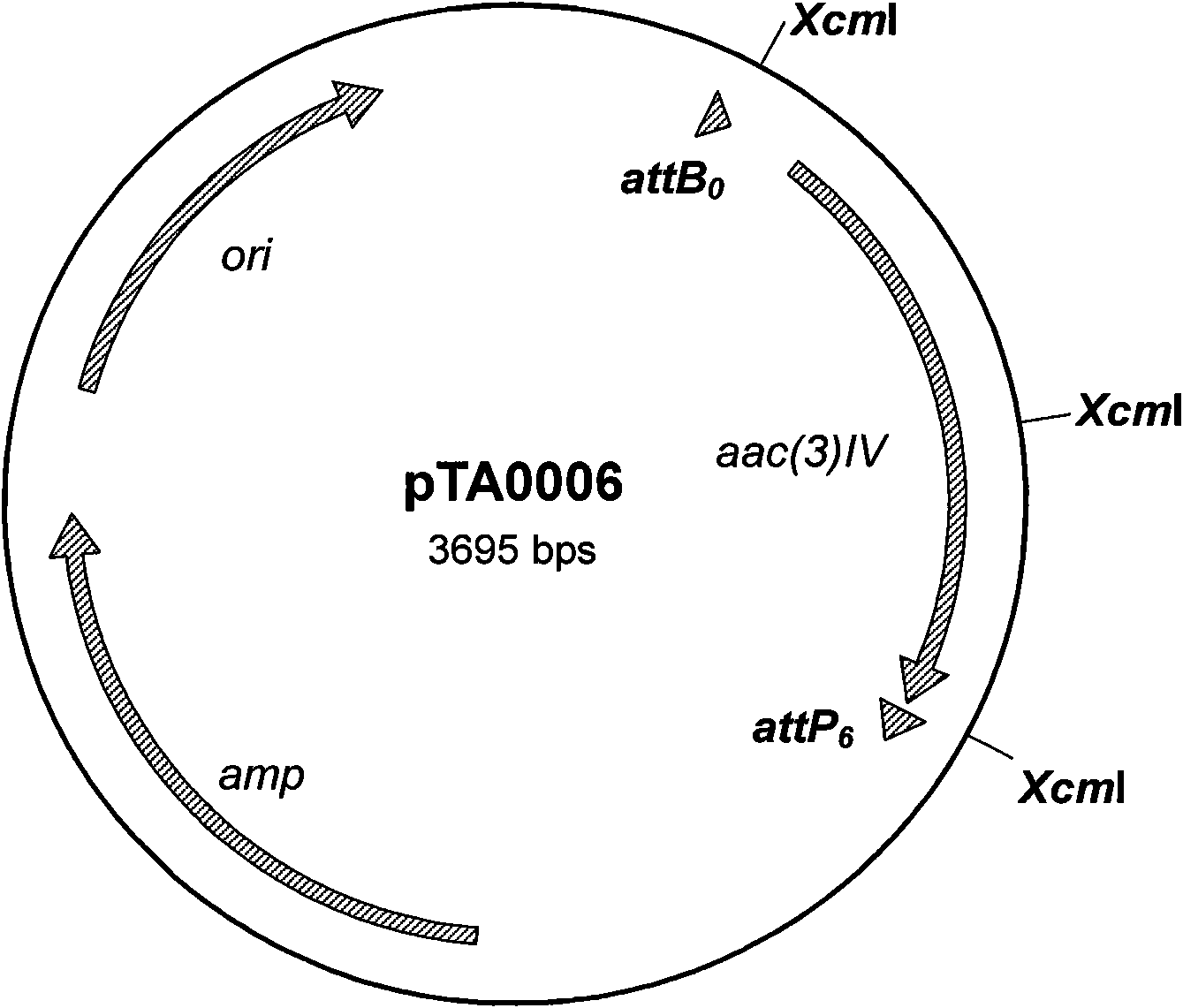
[0036] 1. Acquisition of compatible substrate pairs for φBT1 integrase and determination of recombination efficiency.
[0037] (1) Due to the construction of a series of plasmids for detecting the reaction efficiency of mutant substrates: primers primer 1 and primer 2 are complementary, primer 3 and primer 4 are complementary, primer 5 and primer 6 are complementary, primer 7 and primer 8 are complementary; dissolve the above primers to The final concentration is 20 μM. Take 10 μl each and mix with corresponding complementary primers, then cool and anneal in boiling water. After annealing, double-stranded DNA molecules containing wild-type and mutation sites are obtained, and both ends are respectively digested with EcoRI and HindIII. after the same end. Digest with EcoRI and HindIII (system: appropriate amount of plasmid, 5 μl of NEB “10×buffer”, 5 μl of 10×BSA, 1 μl of restriction endonucleases, ddH 2 (2) Supplement the total volume to 50 μl) plasmid pBC-SK (-), and the dig...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com