Methods of diagnosing chronic cardiac allograft rejection
A graft rejection, allogeneic technique, applied in the field of diagnosing chronic cardiac allograft rejection, can solve the problems of little practicality, no success, difficult tissue evaluation, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0095] Regarding the preparation of monoclonal antibodies against biomarkers, any technique for the production of antibody molecules by continuous cell lines in culture can be used. Such techniques include, but are not limited to, the hybridoma technique originally developed by Kohler and Milstein (1975, Nature 256:495-497), the tripleoma technique (Gustafsson et al., 1991, Hum. Antibodies Hybridomas 2:26- 32), human B-cell hybridoma technology (Kozbor et al., 1983, Immunology Today 4:72) and EBV hybridoma technology for producing human monoclonal antibodies (Cole et al., 1985, see: Monoclonal Antibodies and Cancer Therapy, Alan R. Liss, Inc., pp. 77-96). Human antibodies can be used and can be obtained by using human hybridomas (Cote et al., 1983, Proc. Natl. Acad. Sci. USA 80:2026-2030) or by in vitro transformation of human B cells with EBV virus (Cole et al. et al., 1985, in: Monoclonal Antibodies and Cancer Therapy, Alan R. Liss, Inc., pp. 77-96). Techniques developed f...
Embodiment approach
[0202] T cell atlas of alloreactivity, metabolomics
[0203] T-cell profiling and / or metabolite ("metabolomics") profiling of alloreactivity can be used in combination with genomic and / or proteomic profiling. Small changes in the subject's genome (such as single base changes or polymorphisms) or small changes in genome expression (such as differential gene expression) may lead to rapid responses in the subject's small molecule metabolite profile. Small-molecule metabolites can also respond rapidly to environmental changes, with significant metabolite changes occurring within seconds to minutes of environmental change—conversely, changes in protein or gene expression may take hours or days to manifest. The list of clinical variables indicates several metabolites that can be used to monitor eg cardiovascular disease, obesity or metabolic syndrome - examples include cholesterol, homocysteine, glucose, uric acid, malondialdehyde and ketone bodies. Additional non-limiting examples...
Embodiment 1
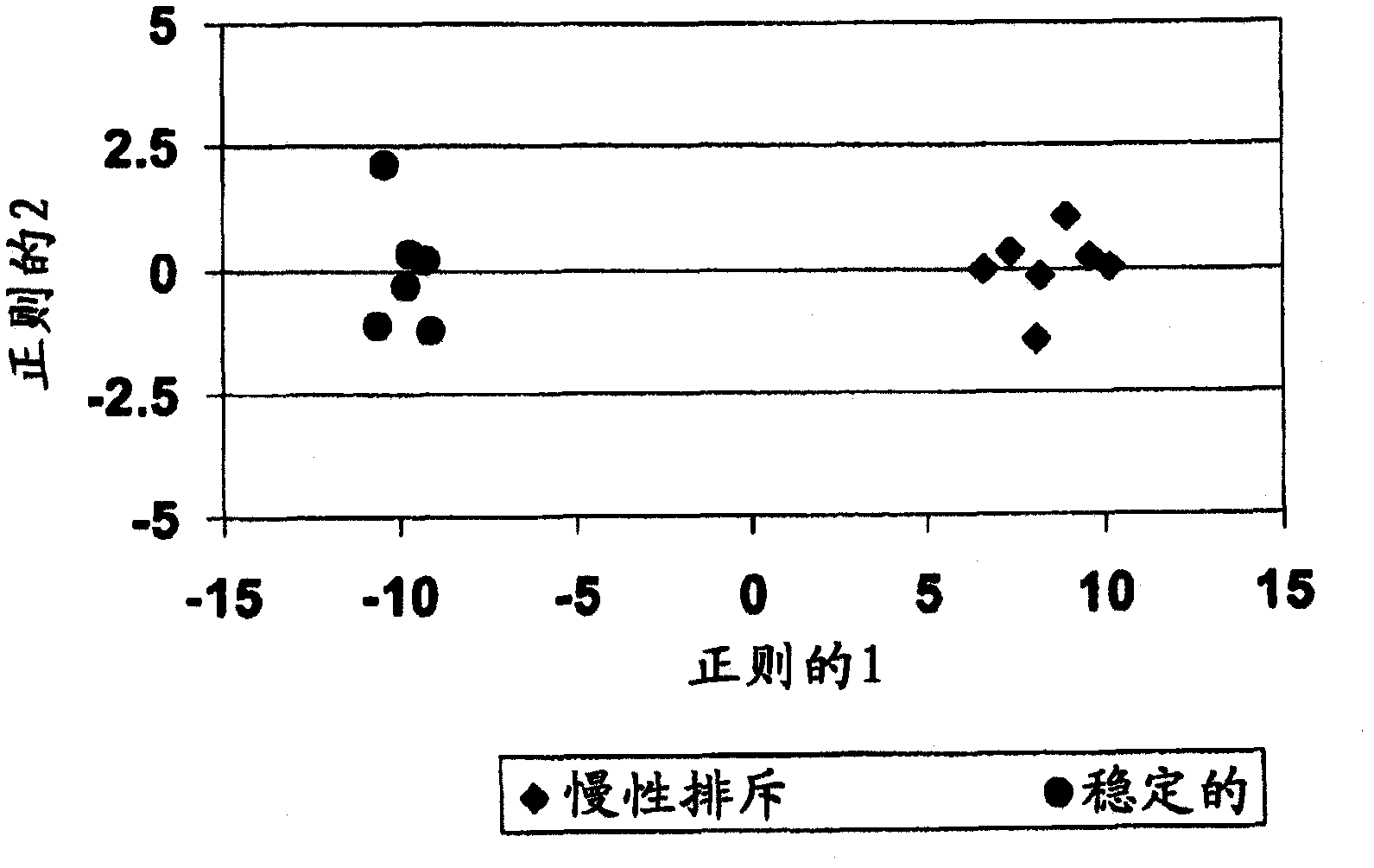
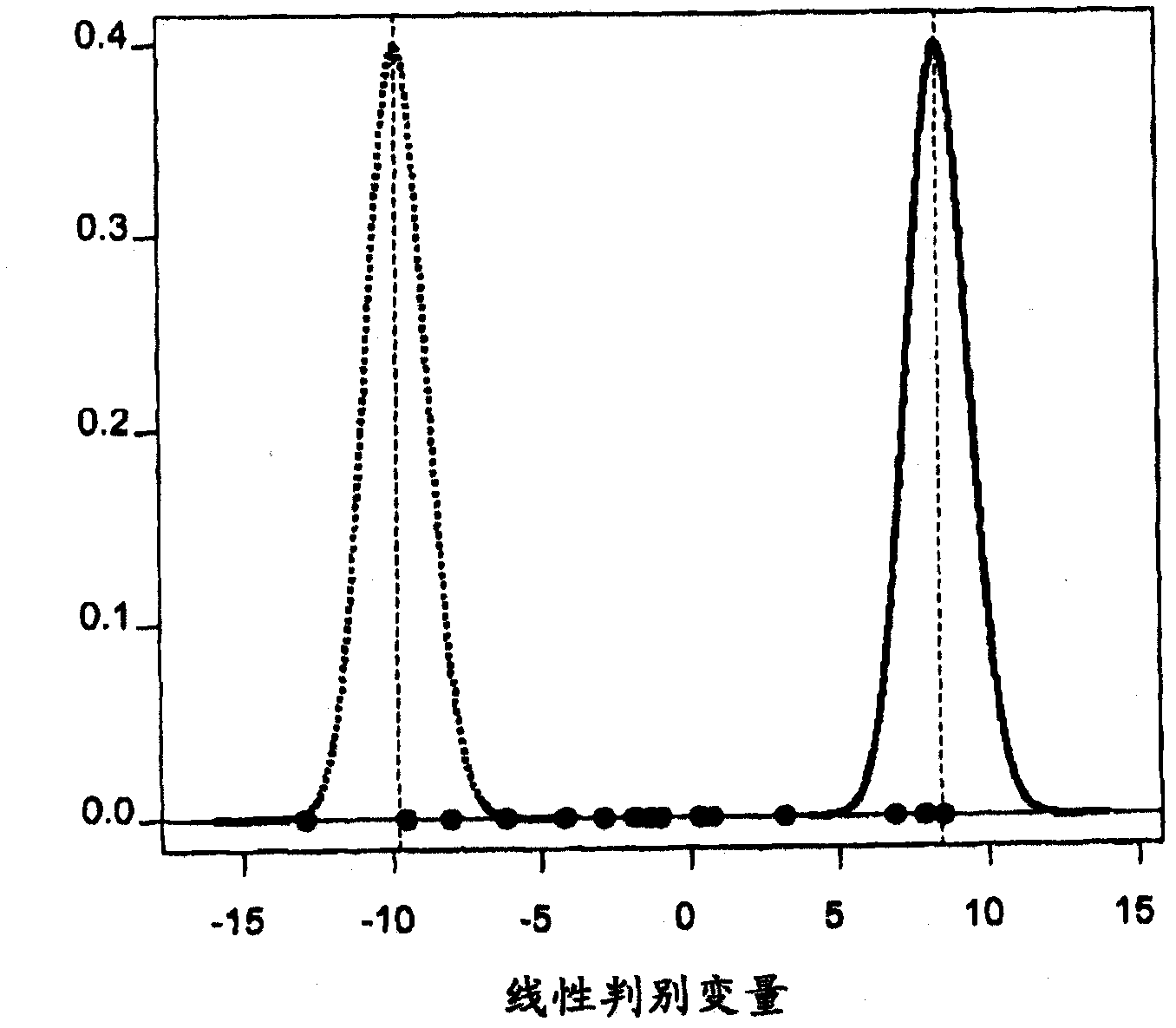
[0252] After normalization and pre-screening, 25,215 probe sets remained and were included in the subsequent analysis using training cohort samples (step 2). Using robust testing, a total of 106 probe sets were identified with FDR Figure 4 ). In addition, overexpression analysis was performed to see the types of biological and molecular processes involved in differentially expressed genes and compared to other genes present on the microarray. Significantly enriched Gene Ontology (GO) terms were identified, those with p-values < 0.05 have been summarized in Table 5.
[0253] Table 5. Statistically significant Gene Ontology terms identified by enrichment analysis of genomic expression profiles (FatiGo). P-value <0.05. The displayed GO terms (biological process and molecular function) are between GO levels 3 and 5.
[0254] process or response
Gene Ontology Terms (GO Terms)
immune response
GO: 0006955
response to biological stimuli
GO: 000...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com