Detection of polyomavirus
A polyoma virus, a technology for detecting samples, applied in the field of detection, can solve problems such as hindering the development of reliable methods for detecting infection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0085] The detection of embodiment 1-polyomavirus
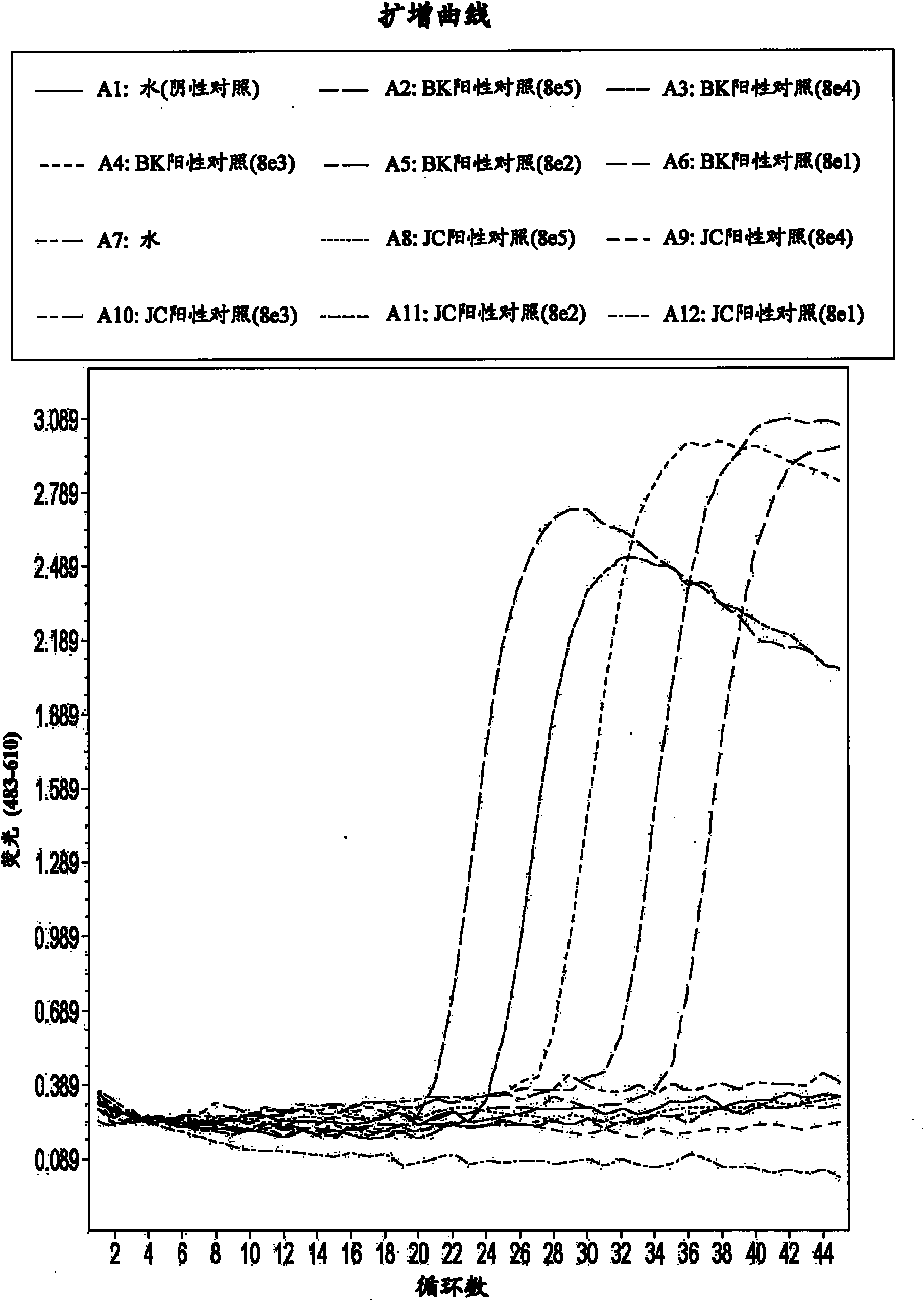
[0086] The real-time amplification assay was performed using primers SEQ ID NO:2 and SEQ ID NO:3 and probes SEQ ID NO:4 and SEQ ID NO:5. The assay includes DNA amplification by polymerase chain reaction (PCR) and real-time detection using a fluorescently labeled donor probe of SEQ ID NO: 4 and an LC610-labeled acceptor probe of SEQ ID NO: 5, which The needles are designed to specifically hybridize to BKV DNA under stringent conditions. Different concentrations of BKV DNA and JCV DNA were detected together with a negative control containing no DNA samples.
[0087] In LightCycler Real-time PCR amplification was performed on a 480 PCR machine (Roche, Basel, Switzerland), and data analysis was performed using LCS480 version 1.2.9.11 software provided by the manufacturer. Reagents from Roche (Basel, Switzerland) were used for all reactions. Each 20-μl PCR reaction contained 1× Fast-Start Hyb Probe mixed master solution (R...
Embodiment 2
[0093] Example 2 - Evaluation of samples containing BKV and JCV
[0094] Samples containing BKV and JCV were evaluated using the general conditions described in Example 1. Well D1 is a negative control that does not contain viral DNA. Microwells D2 to D6 contain 8×10 5 Copy, 8×10 4 Copy, 8×10 3 Copy, 8×10 2 Copy and 8×10 1 Copies of BKV DNA. Microwells D7 to D12 are duplicates of microwells D1 to D6, respectively. Well E1 is a negative control that does not contain viral DNA. Microwells E2 to E6 respectively contained BKV DNA and JCV DNA at a ratio of 1:1 at the following concentrations, E2:10 5 BKV DNA copies and 10 5 JCV DNA copy; E3:10 4 BKV DNA copies and 10 4 JCV DNA copy; E4:10 3 BKV DNA copies and 10 3 JCV DNA copy; E5:10 2 BKV DNA copies and 10 2 JCV DNA copy; E6:10 1 BKV DNA copies and 10 1 A copy of the JCV DNA. Microwells E7 to E12 are duplicate wells of microwells E1 to E6, respectively.
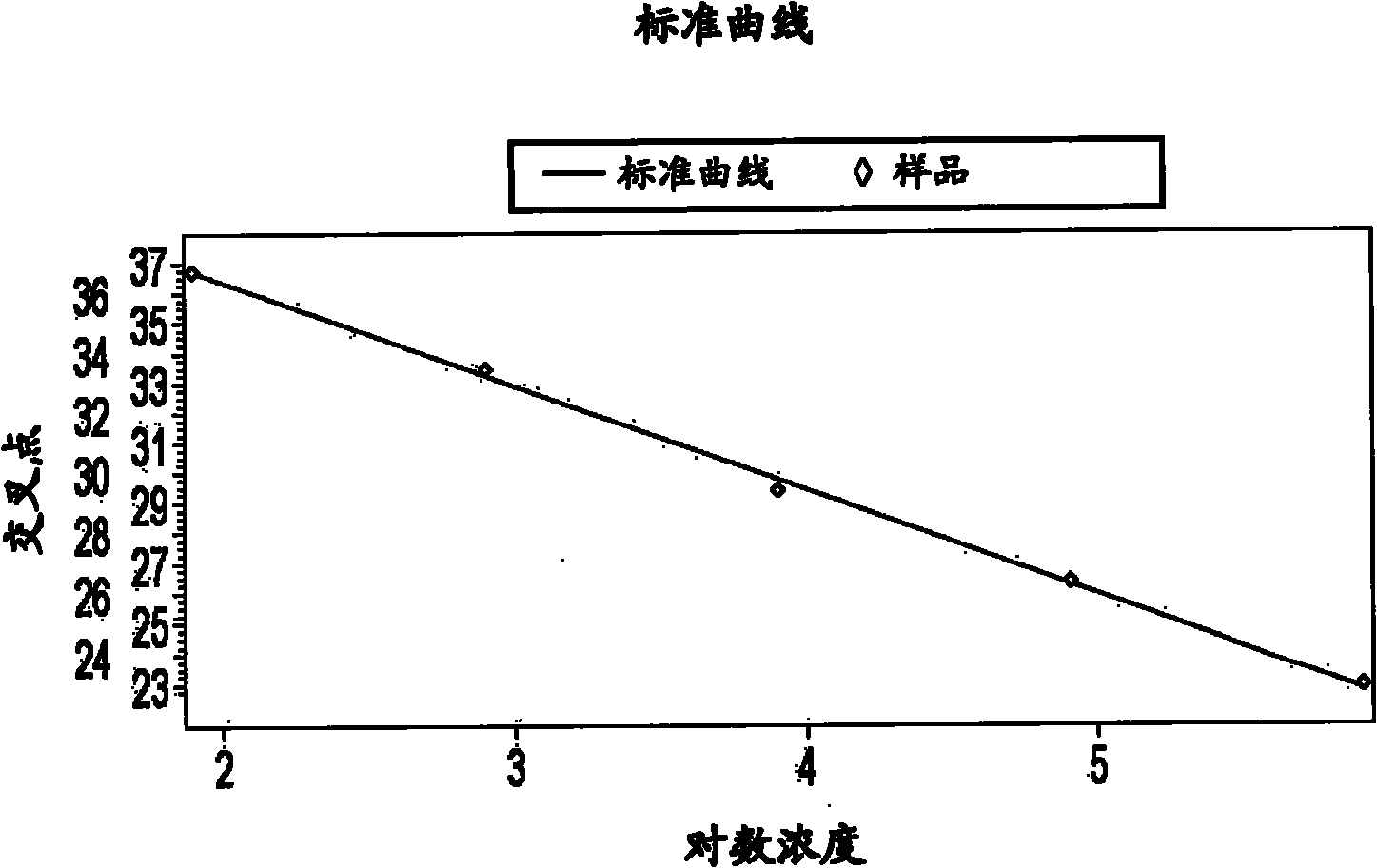
[0095] Figure 4 , Figure 5 as well as Figure 6 Am...
Embodiment 3
[0099] Example 3 - Clinical Trial
[0100] The following primer / probe combinations were used for routine clinical sample testing: combination 1 consisting of primers SEQ ID NO: 6 and SEQ ID NO: 4 and probe sequence SEQ ID NO: 14; combination 1 consisting of primers SEQ ID NO: 6 and SEQ ID NO: 6 and SEQ ID NO: Combination 2 consisting of ID NO: 2 and probe sequence SEQ ID NO: 14; combination 3 consisting of primer BKV_5.2 and SEQ ID NO: 4 and probe sequence SEQ ID NO: 14; and combination 3 consisting of primer BKV_5.2 and Combination 4 consisting of SEQ ID NO: 2 and the probe sequence SEQ ID NO: 14.
[0101] The PCR reaction had a final reaction volume of 40 μl: 10 μl of sample and 30 μl of mixed master mix. For a total volume of 40 μl per sample microwell, the mixed mother liquor composition (30 μl) contained a concentration of 3.125 μM of forward primer, a concentration of 3.125 μM of reverse primer, a concentration of 2.0 to 2.5 μM of the MGB Taqman probe, 20 μl LightCyc...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com