An ubiquitin ligase and the application thereof
A technology of ubiquitin ligase and protein, applied in the field of ubiquitin ligase and its application, can solve the problems that have not been verified or confirmed.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0074] Cloning of the cDNA of embodiment 1, RNF122 gene
[0075] Through bioinformatics analysis, a two-step RT-PCR technique was used to amplify the target gene.
[0076] (1) Search the refseq database of NCBI for human unknown functional gene sequence, obtain the human unknown functional gene sequence, and use the Human_est database to perform sequence correction by BLASTn method, and finally obtain the sequence: SEQID NO: 1 (RNF122 gene, please refer to figure 1 Shown, its coded amino acid sequence please refer to SEQ ID NO: 2 and figure 2 shown). Design specific primers for gene RNF122 according to these sequences:
[0077] Upstream primer (5'-3'): CATTGATGCACCCATTCCAGTG (SEQ ID NO: 3);
[0078] Downstream primer (5'-3'): TTCTCAGGTCTCGGTGTAGCG (SEQ ID NO: 4).
[0079] (2) Using the above primers, select a template from the existing cDNA library and tumor library according to the expression profile of the target gene, and perform initial amplification. The existing li...
Embodiment 2
[0089] Example 2. Construction of pEGFP-N1-RNF122 and pEGFP-N1-RNF122-ΔTM plasmids
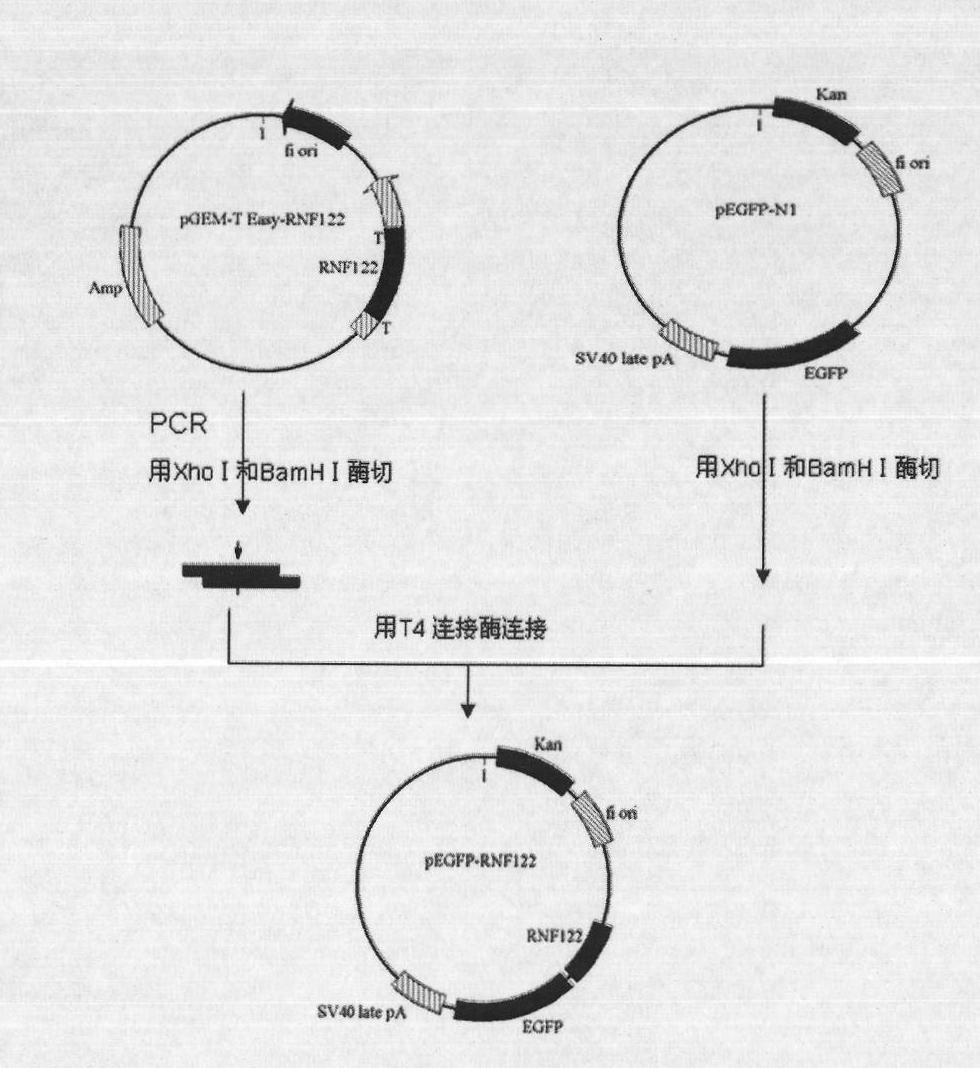
[0090] RNF122 was amplified from pcDB-RNF122 with specific primers (upstream primer (5'-3'): CTCGAGATGCACCCATTCCAGTG (SEQ ID NO: 5); downstream primer (5'-3'): TTCTCCAGGTCTCGGTGTAGCG (SEQ ID NO: 6) fragment, remove the termination code at the same time, and introduce Xho I and BamH I restriction sites, cut the EGFP-N1 vector with corresponding restriction enzymes, connect with T4 ligase, and obtain the fusion expression vector pEGFP-N1-RNF122 of GFP-RNF122 (see image 3 shown). Using primer 5'-CCC CTC GAG AGG ATG AGC AAA CTG CGG AACCA-3' (SEQ ID NO: 7) and primer 5'-CGG GGA TCC ACC ACC AGC TCA TCC AAT AGA-3' (SEQ ID NO: 8) , using pEGFP-N1-RNF122 as a template, PCR amplified the target fragment, after the product was recovered by agarose gel electrophoresis, the recovered fragment was digested with Xho I and BamH I, purified and combined with pEGFP treated with Xho I and BamH I -N1 carrier c...
Embodiment 3
[0091] Example 3, Construction of eukaryotic expression plasmids RNF122C92A-myc and RNF122C95A-myc
[0092] Mutant RNF122 was constructed by PCR-based site-directed mutagenesis. The primers used were P1: 5'-CCGCTC GAG ATG CAC CCA TTC CAG TGG TG-3' (SEQ ID NO: 9), P2: 5'-CGC GGA TCCGGC ACC AGC TCA TCC AAT AGA AT-3' (SEQ ID NO: 10), P3: 5'-AG ACC GCAGTC TGT-3' (SEQ ID NO: 11), P4: 5'-ACA GAC TGC GGT CT-3' (SEQ ID NO: 12), P5: 5'-A GTC CTG GAA GAC TT-3' (SEQ ID NO: 13) and P6: 5'-AA GTCTTC CAG GAC T-3' (SEQ ID NO: 14); where the box marks the introduced mutation site.
[0093] Such as Figure 5 As shown, RNF122C92A-myc uses pcDB-RNF122 as a template, first uses P1 and P4 as primers to amplify, uses P3 and P2 as primers to amplify, and recovers the mixed products of the two; using this as a template, uses P1 and P2 as primers to carry out Amplify, recover and purify, use Xho I and BamH I to cut and recover the product and purify it, then connect it to the pcDB vector, tra...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com