Method for introducing mutated gene, gene having mutation introduced therein, cassette for introducing mutation, vector for introducing mutation, and knock-in non-human mammal
A gene knock-in, mammalian technology, applied in vectors, nucleic acid vectors, genetic engineering, etc., can solve problems such as low probability of homologous recombination, not improving the probability of homologous recombination
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
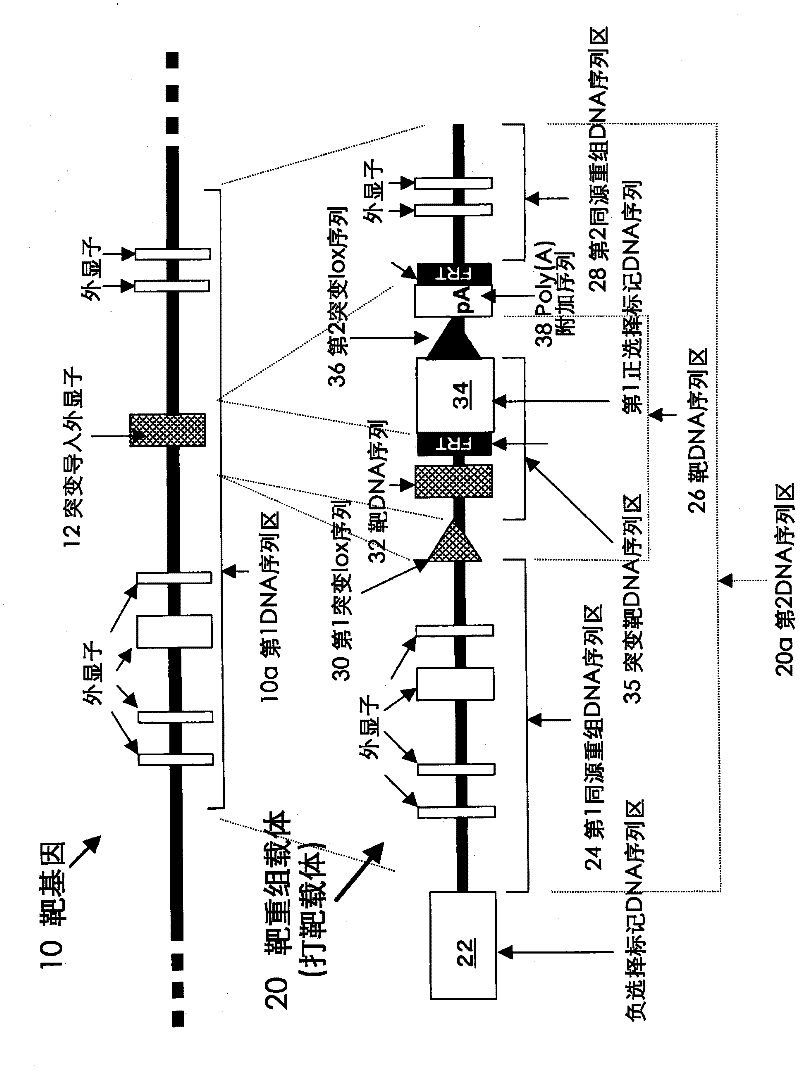
[0156] (1) Sequence structure of target recombinant vector
[0157] The target recombination vector consists of a part derived from a plasmid, a negative selection marker cassette, a partial sequence of the KCNQ2 gene as the first homologous recombination DNA sequence region (long arm region), and lox71 as the first mutant lox sequence from the 5'-end. sequence, a positive selection marker DNA sequence, a lox2272 sequence as the second mutant lox sequence, a second homologous recombination DNA sequence region (short arm region), and a sequence structure composed of a restriction endonuclease site for linearization of the vector composition.
[0158]The target recombination vector (pTgKCNQ2) was constructed based on the pBluescript II SK+ plasmid, with a full-length DNA sequence of 14,164 bp (SEQ ID NO: 5). Its sequence structure is as follows, and Figure 4 Represents a plasmid map.
[0159] Base number 1-673: part derived from pBluescript II SK+
[0160] Base number 674-2...
Embodiment 2
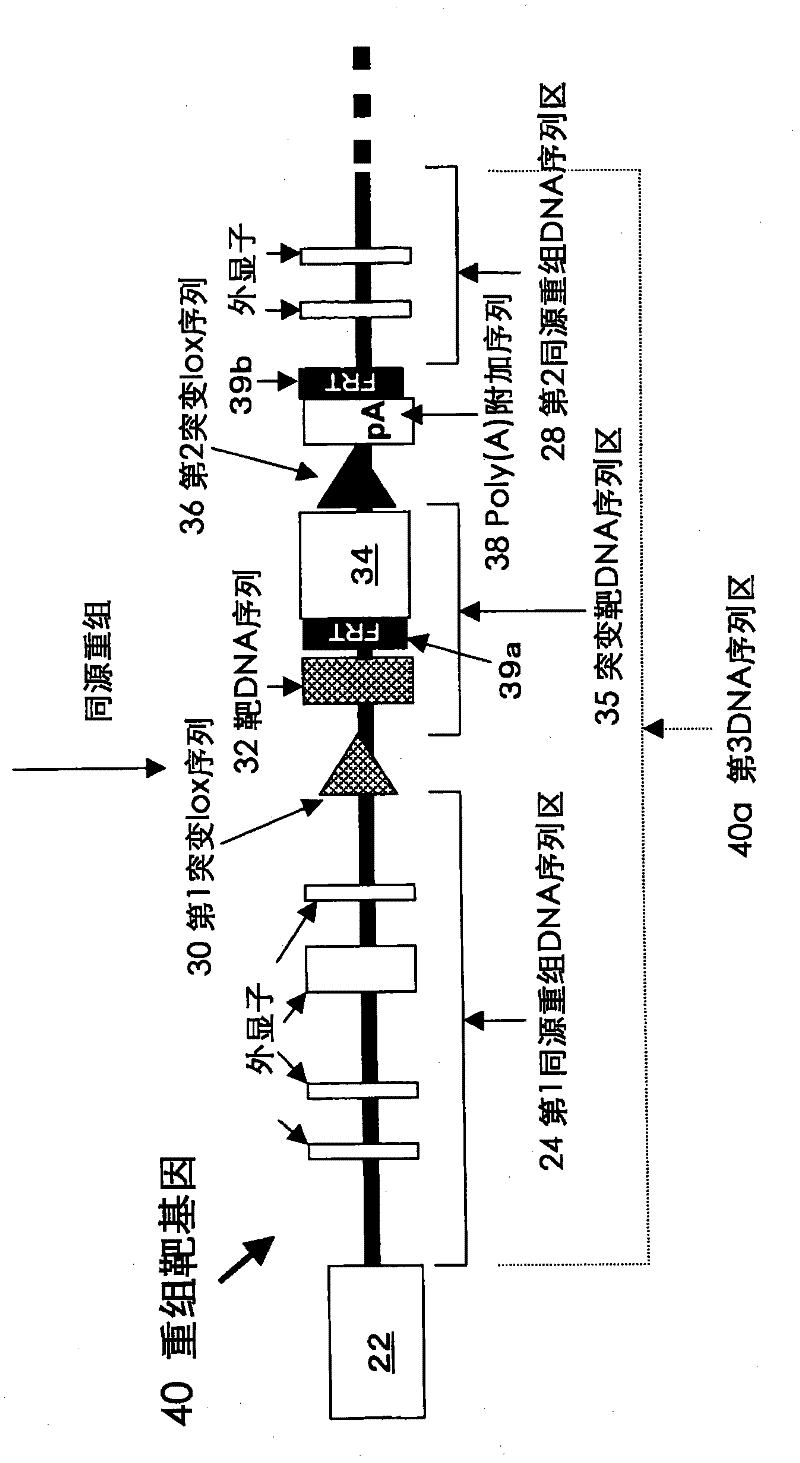
[0200] This example relates to a mutation-introducing vector used in homologous recombination with the target recombination vector prepared in Example 1.
[0201] The KCNQ2 mutation introduction vector (pMtKCNQ2YC and pMtKCNQ2AT) is used to introduce the KCNQ2 gene such as Tyr 284 →Cys or Ala 306 →Two types of vectors for nucleotide replacement of Thr. The loxKR3 sequence is set at the 5' end of the 570bp KCNQ2 gene fragment (exon 6 and before and after) containing any of the above-mentioned arbitrary nucleotide substitutions, and the puromycin resistance gene (PuroR) and lox2272 sequence are set at the 3' end. When these vectors are introduced into the recipient ES cells together with the Cre recombinase expression vector, the region between lox71-lox2272 on the KCNQ2 gene of the recipient ES cells will be efficiently replaced with the mutant KCNQ2 sequence through the action of Cre recombinase . To confer puromycin resistance on the replaced cells, positive selection can ...
Embodiment 3
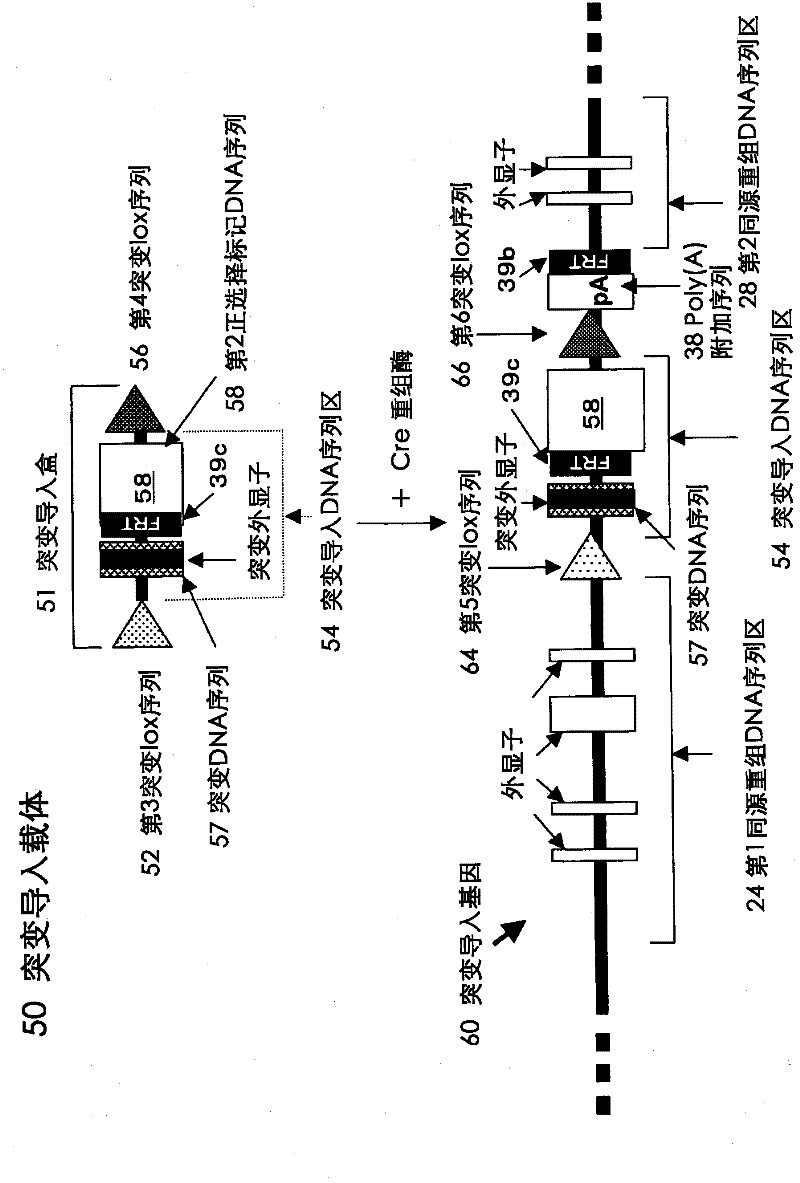
[0230] This example relates to the construction of recipient ES cells capable of introducing mutations into the KCNQ2 gene. The construction of the recipient ES cells is carried out in the following manner (refer to Figure 7 ).
[0231] The mouse ES cells were subcultured in 2 culture dishes, and the semi-confluent (semi-confluent) mouse ES cells (KPTU strain) in a semi-confluent state were detached from the culture dishes by trypsin treatment, and placed in PBS Suspended in medium to a total of 1.6ml. 20 μg of the linearized target recombinant vector prepared in Example 1 was added thereto, and after cooling on ice for 10 minutes, the aliquots were transferred to two cuvettes for electroporation. ), each discharge once under the conditions of 0.8kV and 3.0μF, and carry out the introduction of DNA.
[0232] As described above, the target recombination vector was electroporated to introduce DNA into mouse ES cells, and the above-mentioned sequence factors were introduced in...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com