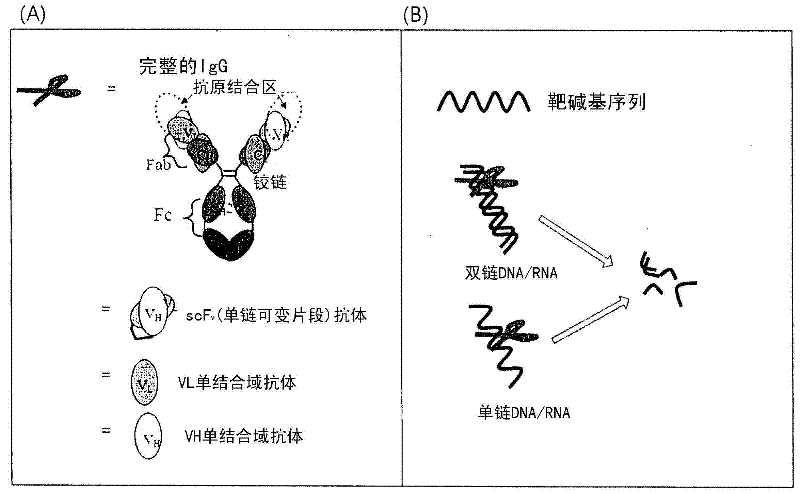
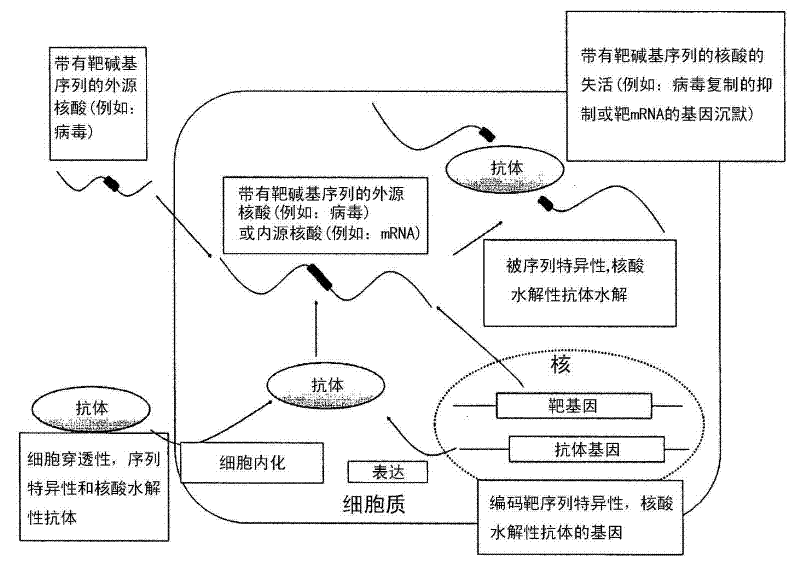
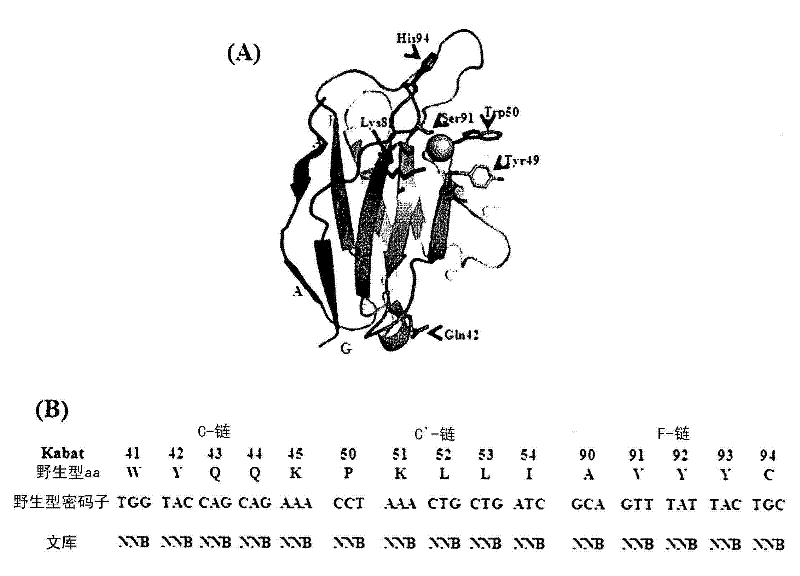
Cell-penetrating, sequence-specific and nucleic acid-hydrolyzing antibody, method for preparing the same and pharmaceutical composition comprising the same
A hydrolyzable and specific technology, applied in chemical instruments and methods, antibodies, antibodies, etc., can solve problems such as limited production
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0060] Example 1 : Design of 3D8 VL 4M Antibody
[0061] 1. Expression of 3D8 VL 4M Antibody on Yeast Cell Surface
[0062] The first step in designing the 3D8 VL antibody to be sequence-specific, nucleolytic antibody is to display the antibody on the surface of yeast cells. The antibody is 3D8 VL 4M, which has better DNA / RNA hydrolysis activity compared with wild type (WT). 3D8 VL 4M has 4 mutants - Q52R, Y55H, W56R and H100A. In order to express the 3D8 VL 4M antibody on the yeast cell surface, the 3D8 VL 4M gene was subcloned from the Escherichia coli expression vector pET23M 3D8 VL 4M into the yeast cell surface display vector pCTCON. To amplify the 3D8 VL4M gene, a primer pair with NheI / BamHI recognition sites was designed. The exact insertion of the 3D8 VL 4M gene in pCTCON was confirmed by base sequencing analysis, and then the recombinant vector was transformed into Saccharomyces cerevisiae EBY100. Transformed colonies were grown in selective SD-CAA medium (20g / L...
Embodiment 2
[0066] Example 2 : Construction of 3D8 VL 4M Antibody Library
[0067] Following the observation that 3D8 VL 4M was expressed at high levels on the surface of yeast cells, a 3D8 VL 4M library was constructed. In order to generate variants that can specifically bind and hydrolyze specific base sequences, the library was constructed based on the template of 3D8 VL 4M. First, the structure of 3D8 VL was analyzed to determine the putative nucleic acid binding site composed of c-, c'- and f-β-strands. It is designed to randomly use degenerate NNB codons (N =A / T / C / G, B=C / G / T) target mutant residues to generate libraries on the surface of yeast cells. Because not all residues of 3D8 VL were mutated, overlap PCR was performed using 4M as a template and mutation primers (some residues of which were mutated) to construct a gene library for yeast surface display. The base sequences of the primers (1F, 2R, 3R, 4F, 5R, 6F, 7R) used for library construction are SEQ ID NOS: 5-11. In th...
Embodiment 3
[0076] Example 3 : Selection of specific variants of the target sequence from the 3D8 VL 4M library
[0077] 1. Screening the 3D8 VL 4M library using competitors
[0078] Two constructed libraries against 5'-biotinylated DNA were screened using MACS and FACS. MACS and FACS screens were performed at high salt concentration (0.3 M) to exclude non-specific binders, which interact with the DNA phosphate backbone through electrostatic interactions. To ensure that selected 3D8 VL variants will specifically bind to a given target sequence, non-biotinylated non-target competitor (DNA) is added to the target substrate. N 18 DNA for Her2 18 competitor. To detect selective binding to G 18 clones, under 0.3M NaCl concentration, the A 18 , T 18 and C 18 Three types of DNA were used as competitors. 5'-biotinylated substrate (G 18 , Her2 18 ) base sequences are used to screen variants specific to the target base sequences, and the target base sequences are respectively represe...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com