Genetic engineering bacteria of gluconobacter oxydans and construction methods thereof
A technology of genetically engineered bacteria and glucose oxidation, applied in the field of genetically engineered bacteria of Gluconobacter oxydans and its construction, can solve the problems of resource and environmental pollution, low vitality, and high raw material prices
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
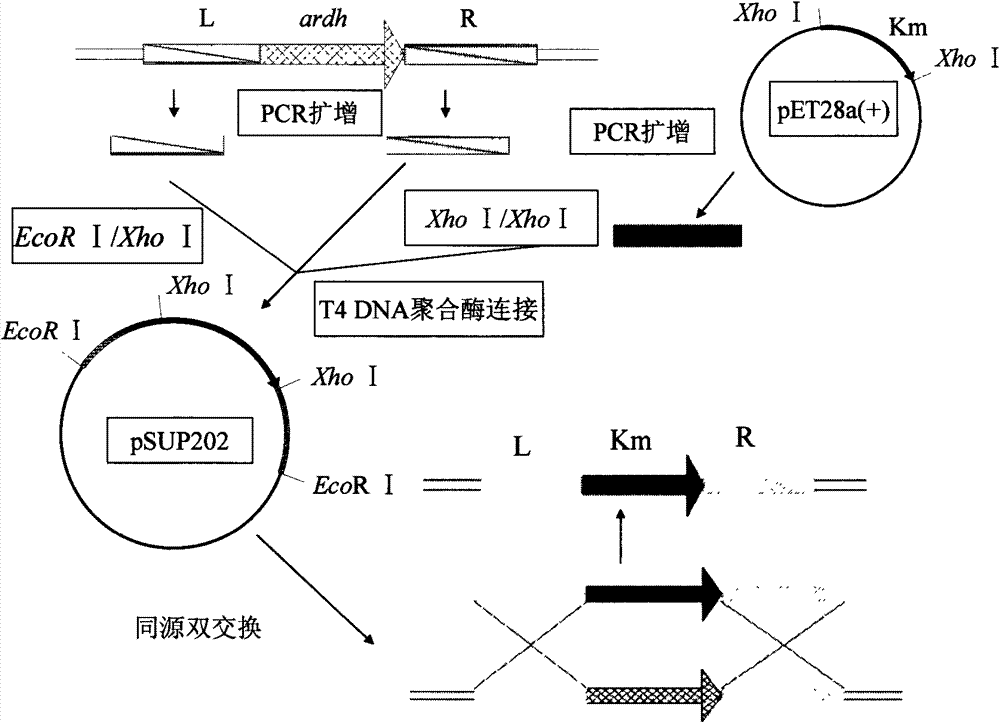
[0080] Example 1: Construction of knockout vector for NADP-arabitol dehydrogenase gene.
[0081] (1) Obtain the upstream s-ardh L and downstream s-ardh R and Km genes of the s-ardh gene.
[0082] Two pairs of primers were designed based on the s-ardh sequence of the Gluconobacter oxydans 621H gene published by GenBank:
[0083] s-ardh L-fwd: 5'-TAT GAATTC CCTCTTGAAAACCTATCATAGC-3'EcoR I,
[0084] s-ardh L-rev: 5'-CTGTTTATGTAAGC CTCGAG AAACTTGAAGTCC-3'Xho I,
[0085] s-ardh R-fwd: 5'-AATAAACAAATAG CTCGAGA AAATGGCCGGGAAG-3'Xho I,
[0086] s-ardh R-rev: 5'-AT GAATTC ATGGCGACTGTCGAACTCAAG-3' EcoR I.
[0087] s-ardh L-fwd and s-ardh R-rev primers were added with the same restriction site EcoR I and protective bases at both ends. s-ardh L-rev and s-ardh R-fwd primers add the same restriction enzyme site Xho I at both ends, and the underlined part is the enzyme site.
[0088] By PCR technology, using CGMCC No.2709 genomic DNA as a template, using s-ardh L-fwd and s-ardhL-r...
Embodiment 2
[0103] Embodiment 2: Obtaining of s-ardh gene knockout mutant
[0104] E.coli JM109 containing the knockout mutant vector pSUP202-s-ardh::Km was used as the donor strain for triparental conjugation, and the recipient strain was CGMCC No.2709. The knockout mutant vector entered CGMCCNo.2709 with the help of the helper bacteria pRK2013, and integrated the target gene fragment containing the Km gene into the chromosome of G.oxydans NH-10. Triparental zygotes were screened by Cefotaxime, Km.
[0105] The donor bacteria E.coli JM109 / pSUP202-s-ardh::Km and the helper bacteria JM109 / pRK2013 were inoculated in LB medium with kanamycin and cultured overnight, and the recipient bacteria G.oxydans were inoculated in G- Ara cultured for 15-20 hours, A 660 Conjugation experiments were carried out when the value was 0.6 to 1.0; the bacterial cells were collected according to the volume ratio of the recipient parent bacteria: helper bacteria: donor bacteria = 3:1:3, that is, the recipient ...
Embodiment 3
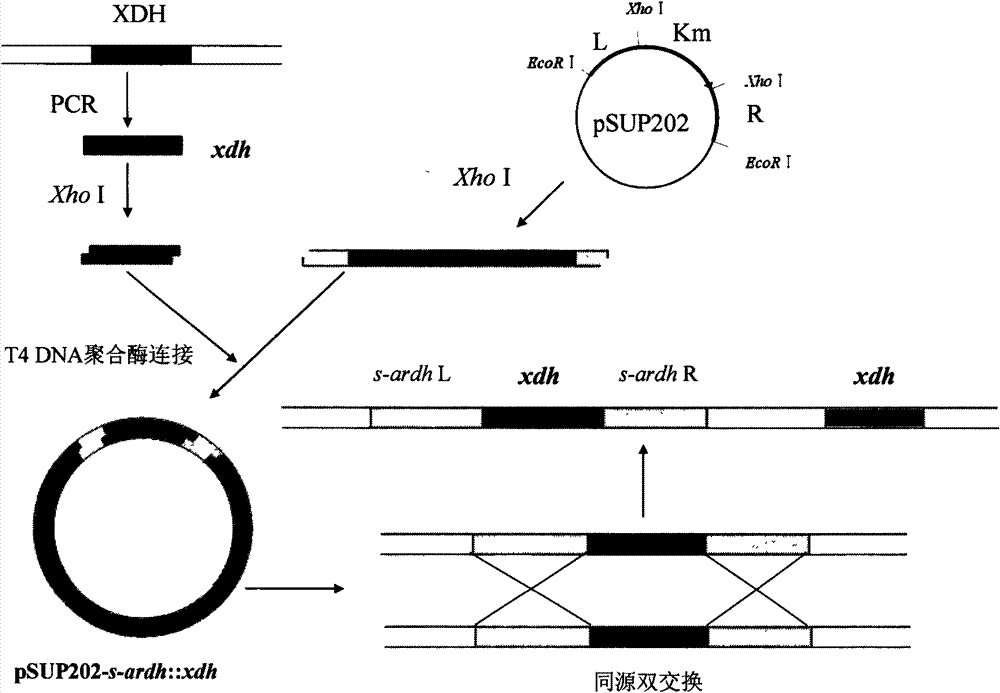
[0107] Example 3: Construction of xylitol dehydrogenase enhancing vector.
[0108] (1) Obtain the xdh gene
[0109] A pair of specific primers were designed based on the xdh sequence of the Gluconobacter oxydans 621H gene published by GenBank. The primers are as follows:
[0110] xdh-fwd:5'-AGG CTCGAG TCGAAGAAGTTTAAG-3'Xho I,
[0111] xdh-rev: 5'-ATT CTCGAG TCAACCGCCAGCAAT-3'XhoI.
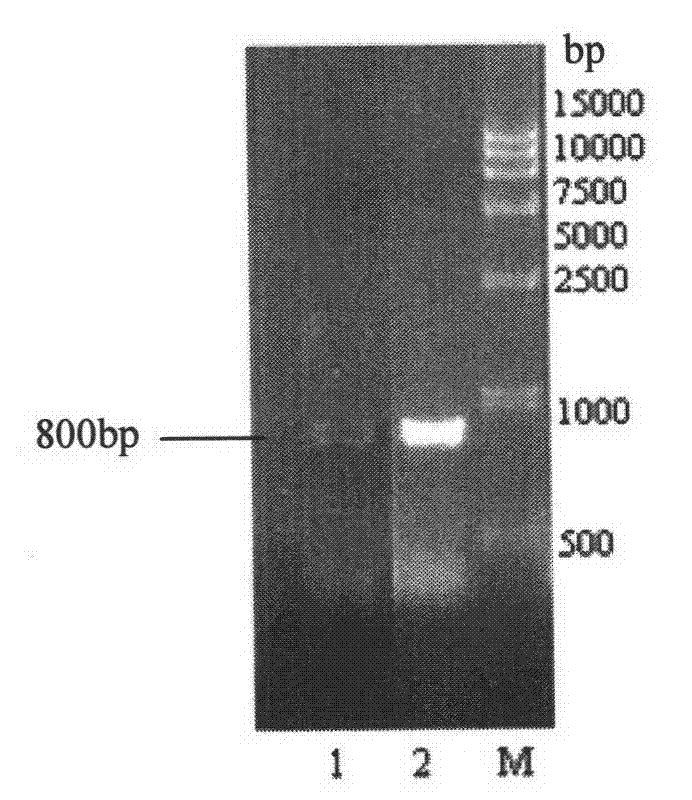
[0112] By PCR technology, with CGMCC No.2709 strain genomic DNA as a template, xdh-fwd and xdh-rev amplified about 800bp xdh sequence ( Figure 9 ). Both ends of xdh-fwd and xdh-rev primers add the same restriction enzyme site Xho I, and the underlined part is the enzyme site.
[0113] xdh gene PCR reaction conditions are: 10×PCR buffer 5μL, Mg 2+ 2 μL, dNTP 5 μL, primers xdh-fwd and xdh-rev 2 μL each, exTaq DNA polymerase 1 μL, genome template 1 μL, add ddH 2 O to a total reaction volume of 50 μL; the PCR reaction was carried out on a PCR machine, and the PCR program was: 94°C pre-denatu...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com