Method for measuring distribution of human cell drug in animal body
A technology of human cells and animal bodies, applied in the biological field, can solve the problems of inability to detect the content of monkey human cells, inability to distinguish human and monkey chromosomal DNA, and narrow application range.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
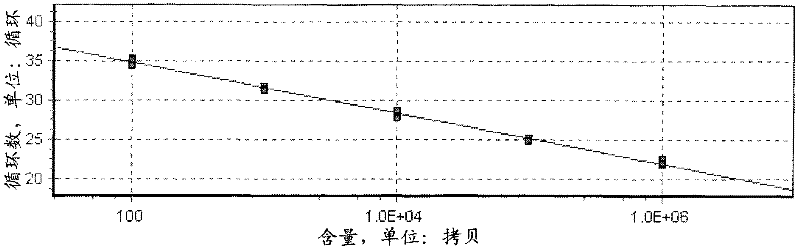
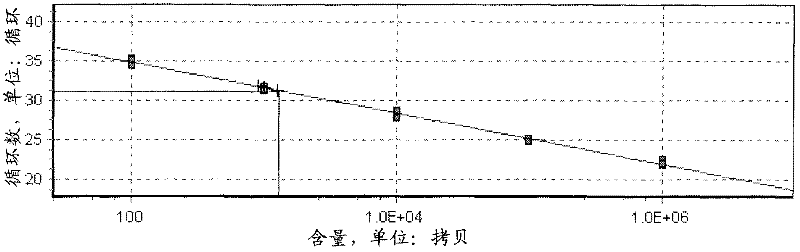
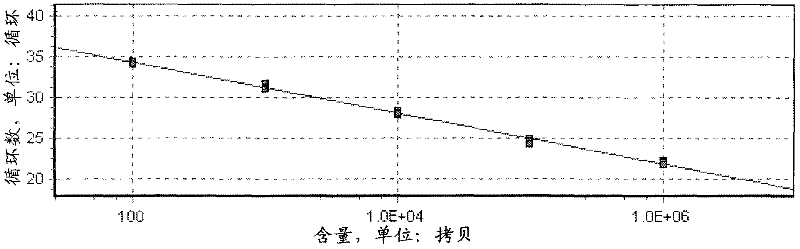
Image
Examples
Embodiment 1
[0058] (1) According to the human specific gene UCMA, select the PCR amplification region (2281-3618), namely SEQ ID NO. 1, and design PCR amplification primers. The primer sequence is as follows.
[0059] Upstream primer: catttagcctgaagtgcctt
[0060] Downstream primer: ccttgtttctcccagcacctctg
[0061] (2) Using conventional tissue total DNA extraction kit (TAKARA, DV811A), extract genomic DNA from whole blood of healthy people, amplify the target gene fragment according to the following reaction conditions, PCR amplification reaction kit (Tiangen, ET101-01 ).
[0062] Reaction system: 0.1ug template, 500nM upstream and downstream primers, add water to 50ul,
[0063] Reaction conditions: 95°C pre-denaturation 10min; 95°C denaturation 15sec, 50°C annealing 15sec, 72°C extension 90sec, 30 cycles; 72°C extension 5min;
[0064] (3) Operate according to the pMD19-T (TAKARA, VT202-01) instruction, connect the target gene fragment into the pMD19-T vector to construct a standard plasmid, and d...
Embodiment 2
[0078] (1) According to the human specific gene UCMA, select the PCR amplification region (10081-10740), namely SEQ ID NO. 3, and design PCR amplification primers. The primer sequence is as follows.
[0079] Upstream primer: ggctcgagcaatcctcccaa
[0080] Downstream primer: ctgcaccccagcctgggtga
[0081] (2) Using conventional tissue total DNA extraction kit (TAKARA, DV811A), extract genomic DNA from whole blood of healthy people, amplify the target gene fragment according to the following reaction conditions, PCR amplification reaction kit (Tiangen, ET101-01 ).
[0082] Reaction system: 0.1ug template, 200nM upstream and downstream primers, add water to 50ul,
[0083] Reaction conditions: 95°C pre-denaturation 10min; 95°C denaturation 15sec, 52°C annealing 15sec, 72°C extension 90sec, 35 cycles; 72°C extension 7min.
[0084] (3) Operate according to the pMD19-T (TAKARA, VT202-01) instruction, connect the target gene fragment into the pMD19-T vector to construct a standard plasmid, and de...
Embodiment 3
[0098] (1) According to the human specific gene UCMA, select the PCR amplification region (11281-11880), namely SEQ ID NO. 5, and design PCR amplification primers. The primer sequence is as follows.
[0099] Upstream primer: gtttcaccatgttggccaagtta
[0100] Downstream primer: tgaccttcgcccatctgggc
[0101] (2) Using conventional tissue total DNA extraction kit, extract genomic DNA from whole blood of healthy people, amplify target gene fragments according to the following reaction conditions, PCR amplification reaction kit (Tiangen, ET101-01);
[0102] Reaction system: 0.1ug template, 300nM upstream and downstream primers, add water to 50ul,
[0103] Reaction conditions: pre-denaturation at 94°C for 10 minutes; denaturation at 94°C for 15 seconds, annealing at 55°C for 15 seconds, extension at 72°C for 90 seconds, 30 cycles; extension at 72°C for 7 minutes.
[0104] (3) Operate according to the pMD19-T (TAKARA, VT202-01) instruction, connect the target gene fragment into the pMD19-T vecto...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com