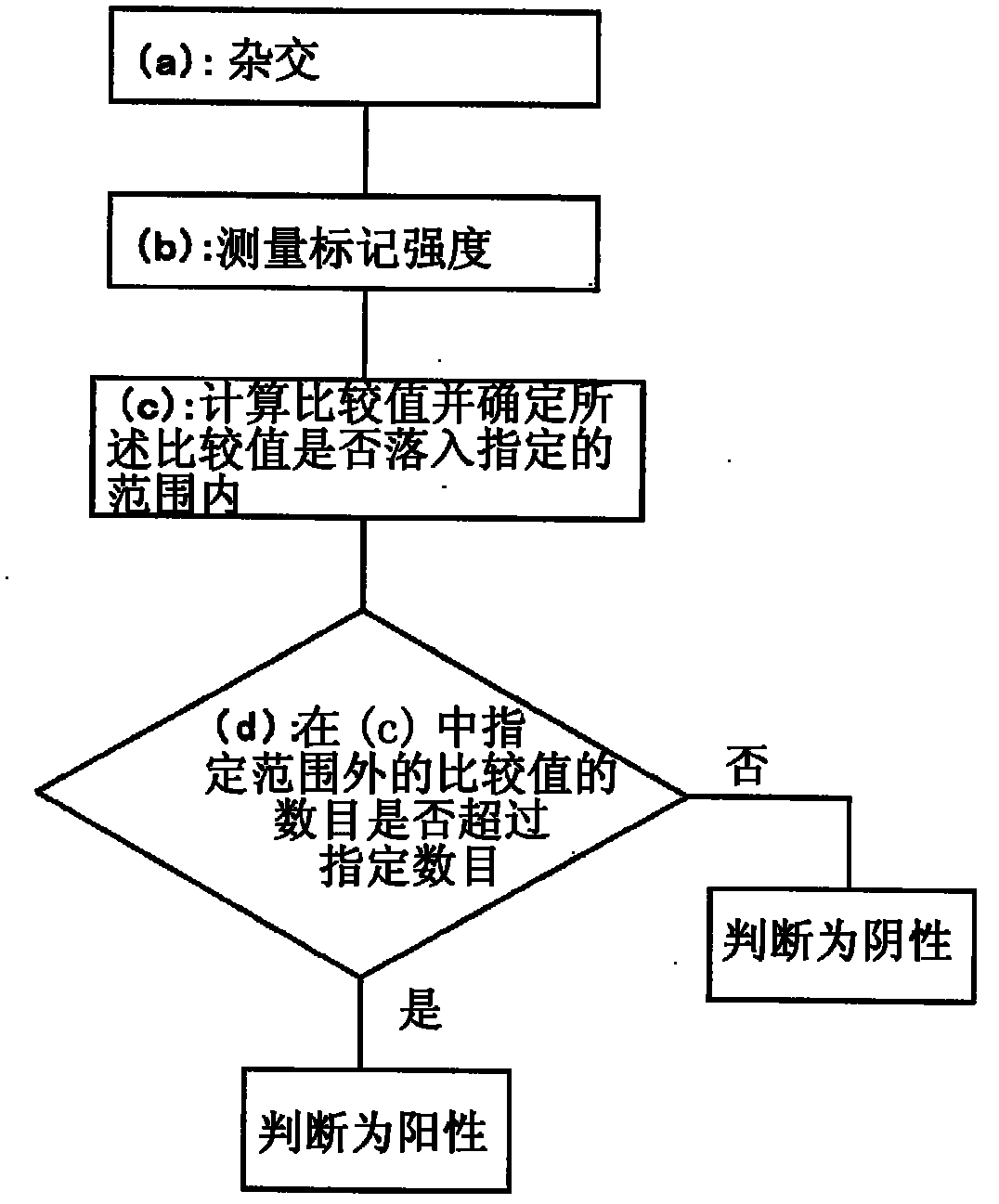
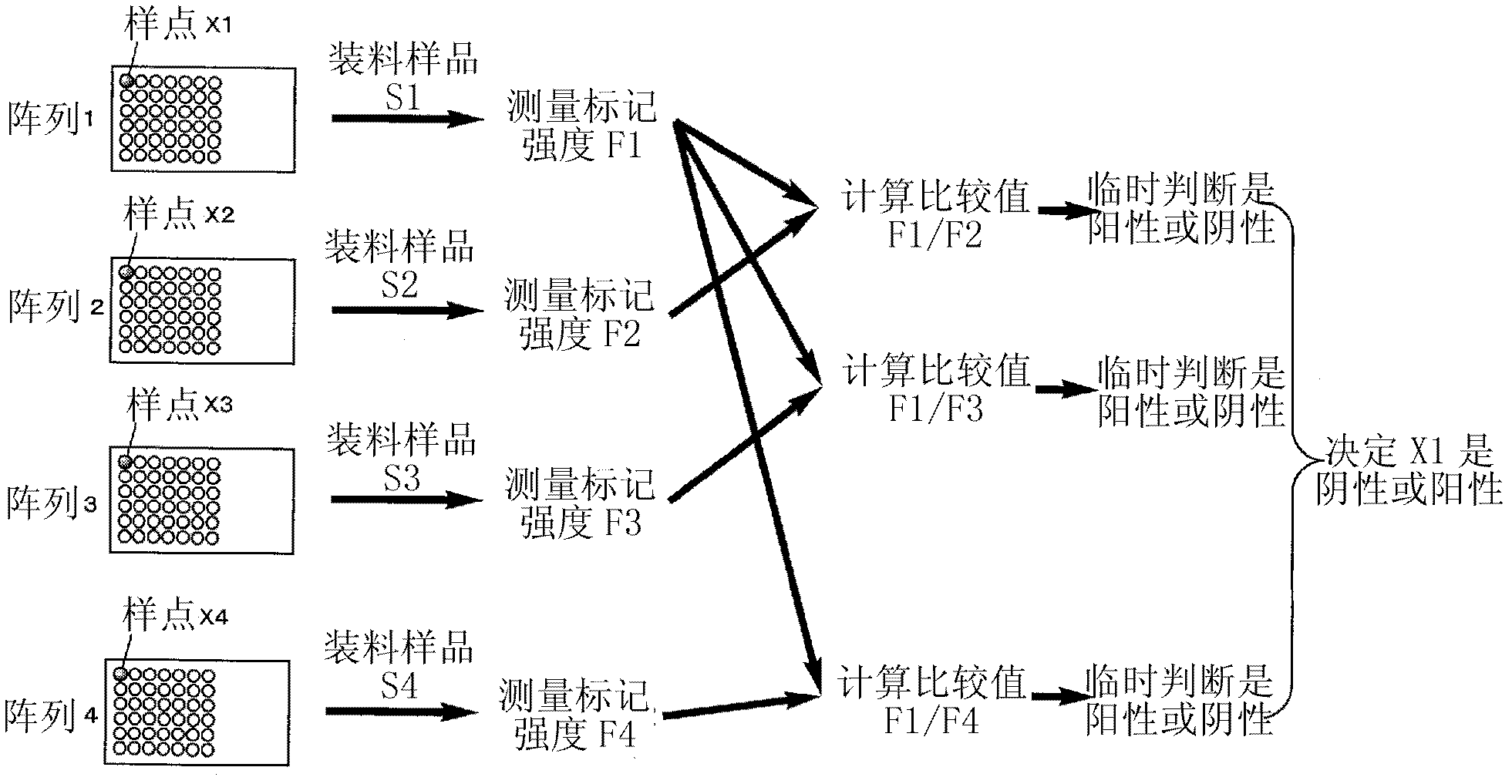
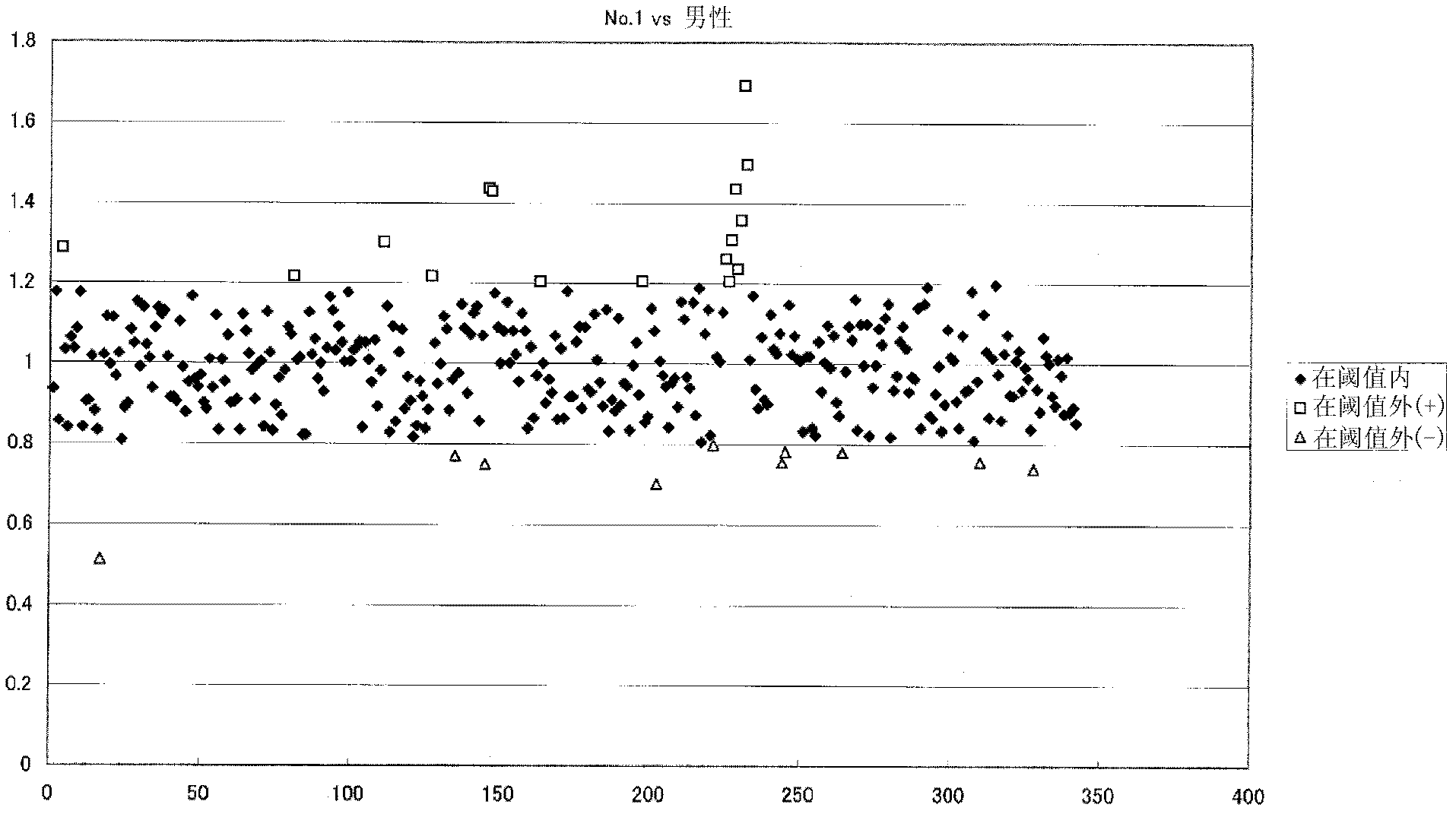
Method For Analyzing Nucleic Acid Mutation Using Array Comparative Genomic Hybridization Technique
A hybridization technology, genome technology, applied in the field of analysis of nucleic acid mutations using array comparative genomic hybridization technology, can solve problems such as inability to make a diagnosis, and achieve the effect of improving reliability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0243] 1. Preparation of Labeled Sample Nucleic Acids
[0244] Put 3 μL (0.75 μg) Human Genomic DNA male (human male genomic DNA) (manufactured by Promega Company, item number G152; hereinafter referred to as male genomic DNA) into the microtube, 8 μL water (distilled water without DNAse, RNase, GIBCO), 20 μL of 2.5×Random Primers Solution (Random Primer Solution) (Invitrogen Company) for the labeling system of Array CGH (BioPrime (registered trademark) Array CGH Genomic Labeling System (Array CGH Genomic Labeling System) (Invitrogen)), then Heat treatment was performed at 95°C for 5 minutes using a block incubator and allowed to stand at 37°C for 15 minutes.
[0245] To this was added 5 μL of 10×dCTP Nucleotide Mix (Nucleotide Mixture) (Invitrogen Company) of the above labeling system, 3 μL Cy3-dCTP Bulk Pack 250 nmol (produced by GE Healthcare Bioscience Company), 1 μL BioPrime (registered trademark) Array CGH Genomic Labeling System (Array CGH Genome Labeling System) Exo-K...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com