Production method for genetically modified plant cells
A technology of transgenic plants and plant cells, applied in the fields of botanical equipment and methods, biochemical equipment and methods, plant products, etc., which can solve problems such as ambiguity and unreported possibility of mutation introduction efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
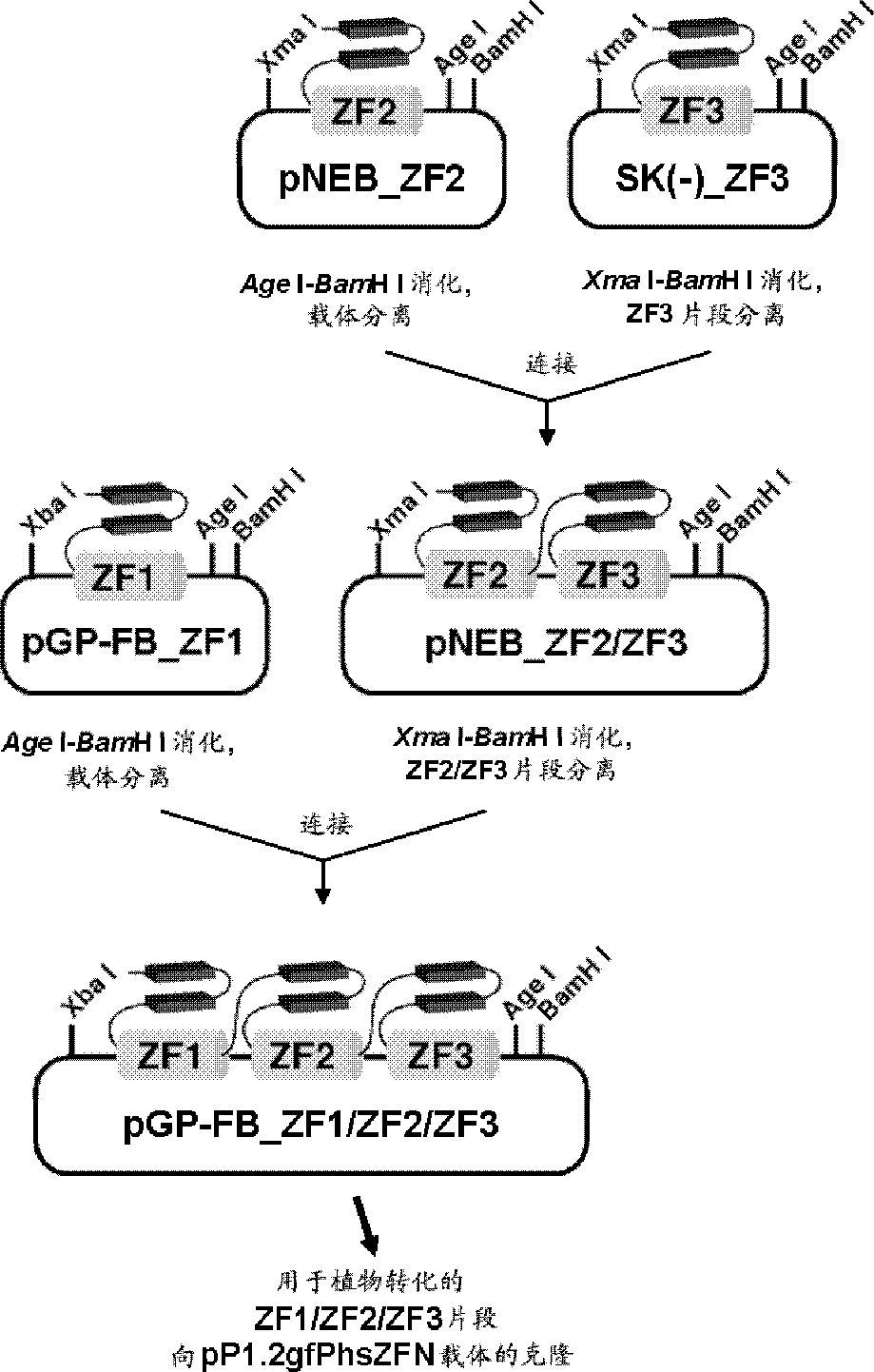
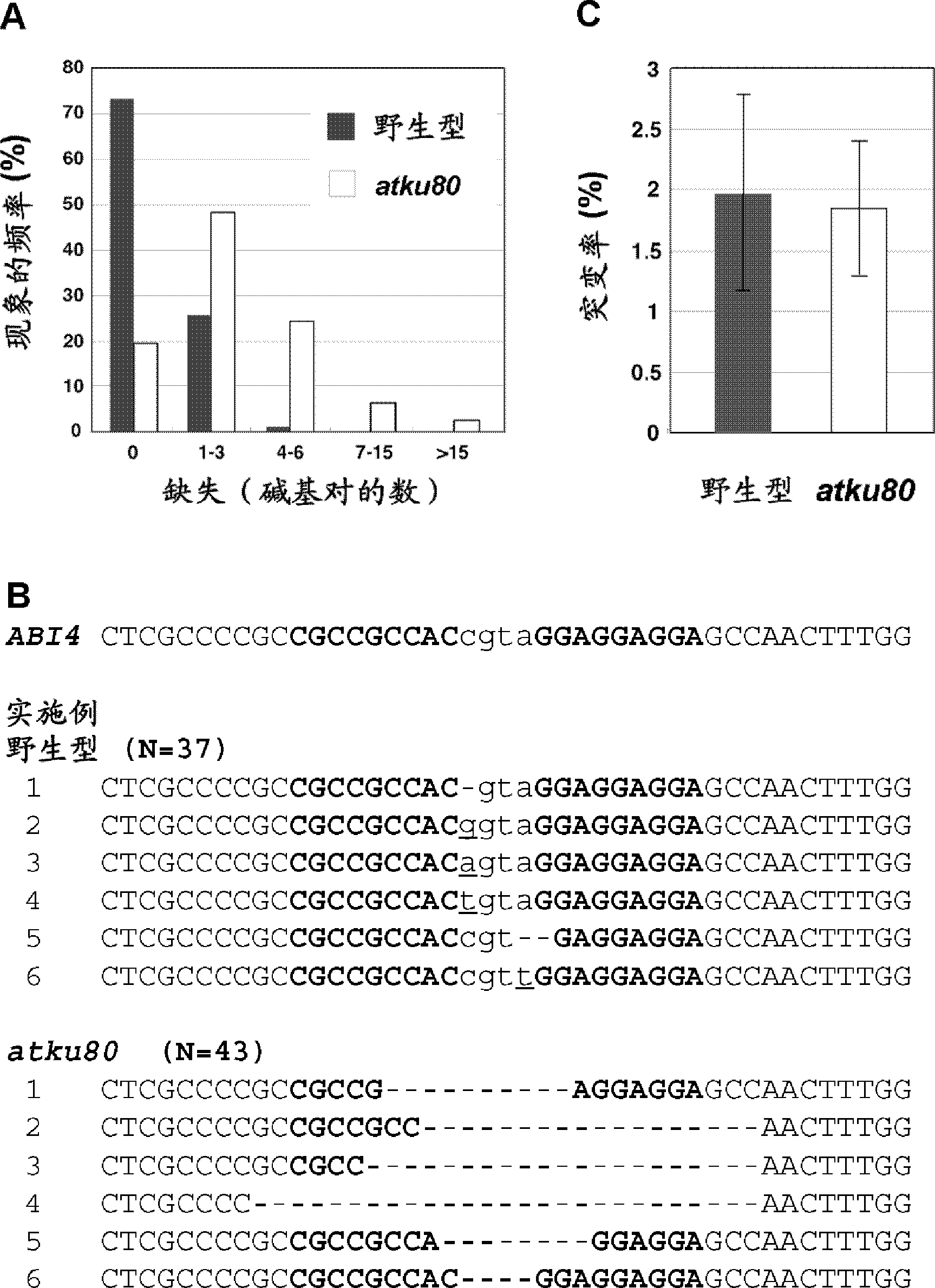
[0099] (Example 1) Introducing a mutation into the ABI4 gene of Arabidopsis using zinc finger nuclease and the effect of the Ku80 deletion of the introduced mutation
[0100] To demonstrate zinc finger-mediated site-directed mutagenesis in Arabidopsis, the inventors chose the ABI4 gene. The present inventors found a complete consensus target site of zinc finger nuclease in ABI4 [5'-NNCNNNNNNNNNNNGNNNGNN-3' (N=A, C, G and T) / SEQ ID NO: 5].
[0101] In this example, in the construction of the modules for the construction of zinc finger nucleases, some changes were made to the method reported by Wright et al. (Wright DA, ea al. Nat Protoc 1:1637-1652, 2006). Methods. For ZF-AAA (with GGA GGA GGA as the target), the amino acid sequences near the specificity determining residues refer to 1 "QRAHLER / SEQ ID NO. 6", 2 "QSGHLQR / SEQ ID NO. 7" and 3 "QRAHLER / SEQ ID NO. 8", for ZF-TCC (with GTG GCG GCG target), it is finger 1 "RSDALTR / sequence number 9", finger 2 "RSDDLQR / serial number ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com