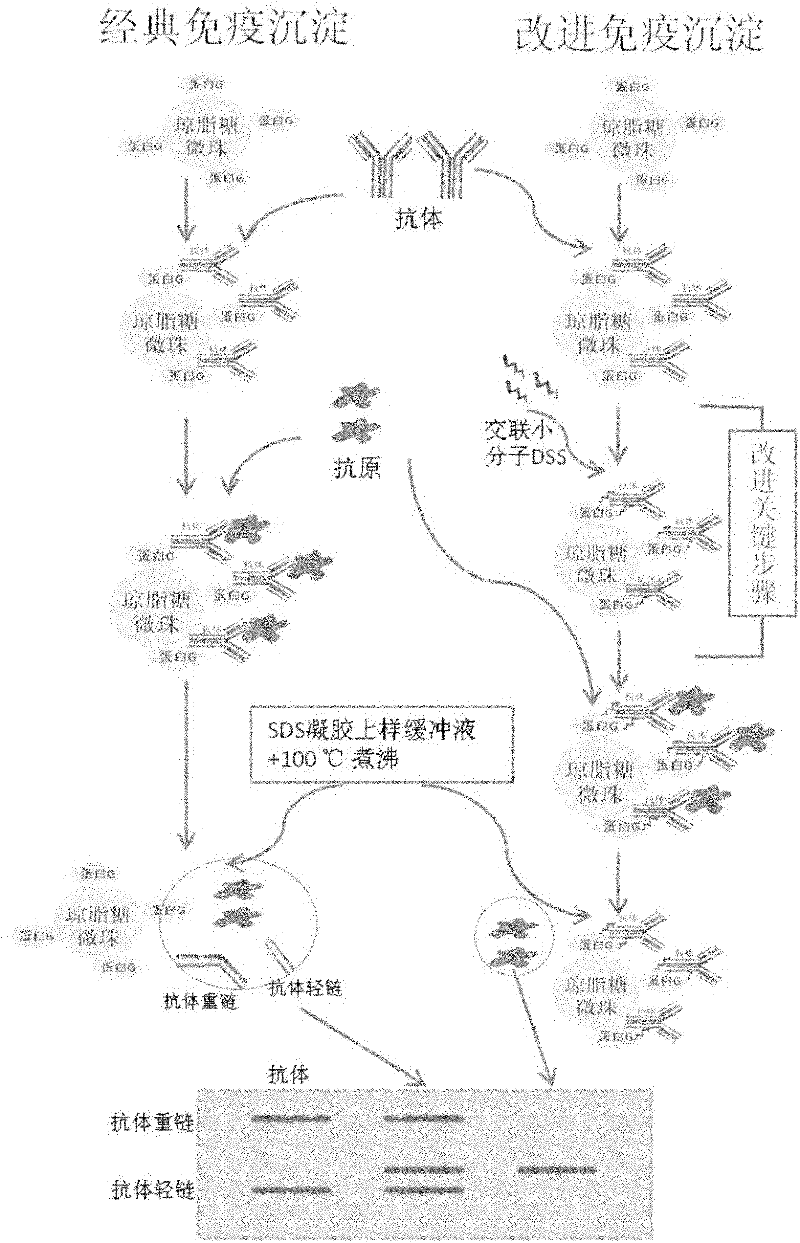
Improved co-immunoprecipitation technical method
A co-immunoprecipitation and protein technology, applied in the field of optimized co-immunoprecipitation technology, can solve the problem of containing target protein and antibody
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0032] Example 1: Comparison between the improved co-immunoprecipitation method and the traditional IP method in identifying the interaction between AIF and actin
[0033] The steps are as follows:
[0034] Extraction of total intracellular protein:
[0035] 1.1 Use a petri dish with a diameter of 10 cm to culture the cells of the esophageal cancer cell line KYSE140 (a human esophageal cancer cell line from Japan, donated by Professor Shimada Y, Kyoto University), in the PRM1640 medium containing 10% fetal bovine serum, Cultivate for 3 days, and when the confluence of the cells reaches 90%, wash the cells with 4°C pre-cooled 1×PBS, 5ml / time×2 times. After washing, add pre-cooled 0.6ml cell lysate to the cell culture dish, the ingredients include 50mM Tris, pH7.4, 150mM sodium chloride, 1% triton-100 (membrane permeabilizer), 0.1 %SDS, and quickly scrape the cells with a cell scraper and collect them in a 15ml centrifuge tube. Sonication on ice (sonication for 10 seconds, wi...
Embodiment 2
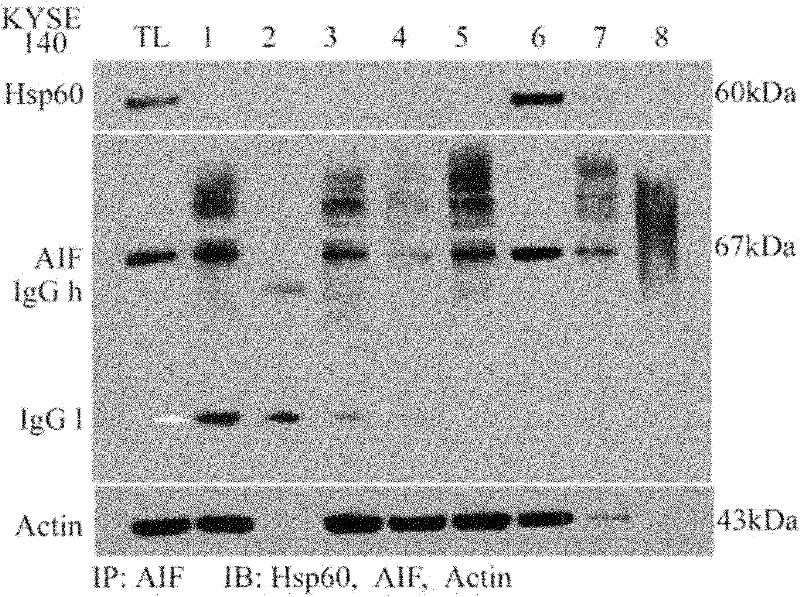
[0054] Example 2: Determining Optimal Antibody and Protein G Optimal Crosslinking Time
[0055] All materials and methods are the same as in Example 1, wherein only the cross-linking time has been changed, the incubation time of step 2.3 in Example 1 is changed to: 50min, 60min and 90min, the results in figure 2 Lanes 3-5 are shown.
[0056] In step 3.4, the samples on SDS electrophoresis include the protein solution of KYSE140 whole cell lysis, the supernatant obtained from the classic IP experiment, the cross-linked 50min, 60min and 90min supernatant obtained from the improved IP experiment, and the eluted supernatant of the IP experiment After SDS electrophoresis and transmembrane transfer, conventional immunoblotting was performed using anti-AIF antibody, anti-Actin antibody and anti-heat shock protein 60 (HSP60) antibody.
[0057] The result of embodiment 2:
[0058] The total protein of the esophageal cancer cell line KYSE140 cells was subjected to the classic co-immu...
Embodiment 3
[0062] Example 3: Using the improved co-immunoprecipitation technique to identify the interaction between RIP3 protein and ENO1 The steps are as follows:
[0063] 1. Extraction of total intracellular protein:
[0064] 1.1 Use two petri dishes with a diameter of 10 cm to culture the cells of the esophageal cancer cell line KYSE140, a human esophageal cancer cell line from Japan, in the PRM1640 medium containing 10% fetal bovine serum, culture for 1 day, and wait for the fusion of the cells up to 80% degree. One plate of cells was treated with 10uM anticancer drug cisplatin for 24 hours (B), and the other plate was not treated as a control (A). Wash the cells with 4°C pre-cooled 1×PBS, 5ml / time×2 times. After washing, add pre-cooled 0.6ml cell lysate to the cell culture dish, the ingredients include 50mM Tris, pH7.4, 150mM sodium chloride, 1% triton-100 (membrane permeabilizer), 0.1 %SDS, and quickly scrape the cells with a cell scraper and collect them into a 1.5ml centrifug...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com