Cross-species-specific psmaxcd3 bispecific single chain antibody
A single-chain antibody and bispecific technology, applied in the direction of antibody, anti-receptor/cell surface antigen/cell surface determinant immunoglobulin, specific peptide, etc., can solve the problems of increasing the risk of prostate cancer and achieve maximum safety , good safety effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0297] Example 1 Generation and characterization of PSMA and CD3 cross-species specific bispecific single chain antibody molecules
[0298] 1.1 Cloning and expression of human PSMA antigen on CHO cells
[0299] The sequence of the human PSMA antigen ('AY101595', Homo sapiens prostate specific membrane antigen mRNA, complete cds, was synthesized by gene synthesis according to standard operating procedures, National Center for Biotechnology Information, http: / / www.ncbi.nlm.nih .gov / entrez) for obtaining synthetic molecules. Gene synthesis fragments were also designed to contain Kozak sites for eukaryotic expression of the construct and restriction sites at the beginning and end of the DNA. The introduced restriction sites (XbaI at the 5' end and SalI at the 3' end) were used during the cloning step in Mack et al. (Mack M et al., ProcNatl Acad Sci USA 1995;92:7021-5 and Raum et al. Human Cancer Immunol Immunother (2001) 50 (3)) described in the expression plasmid called pEFDH...
Embodiment 2
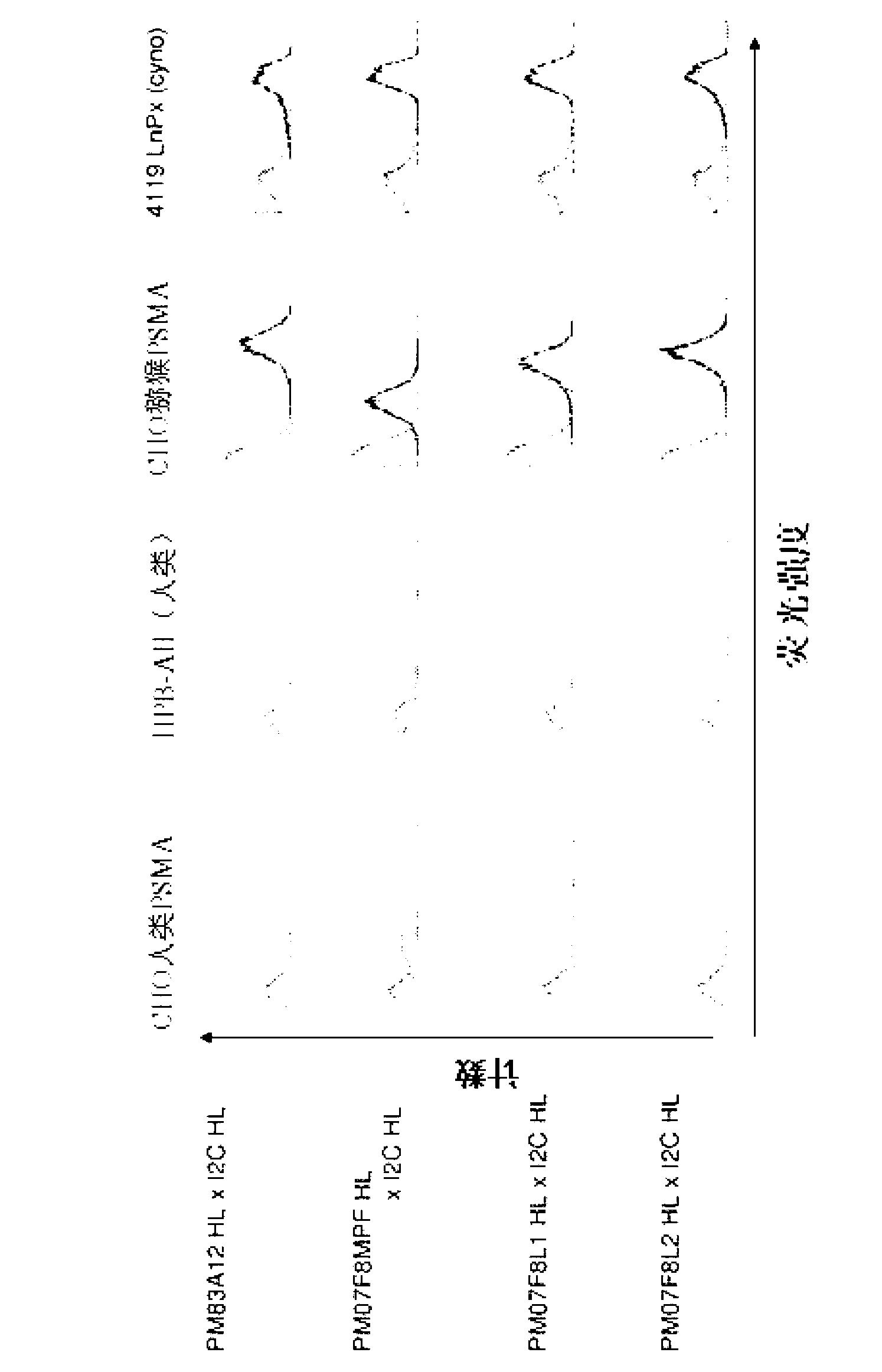
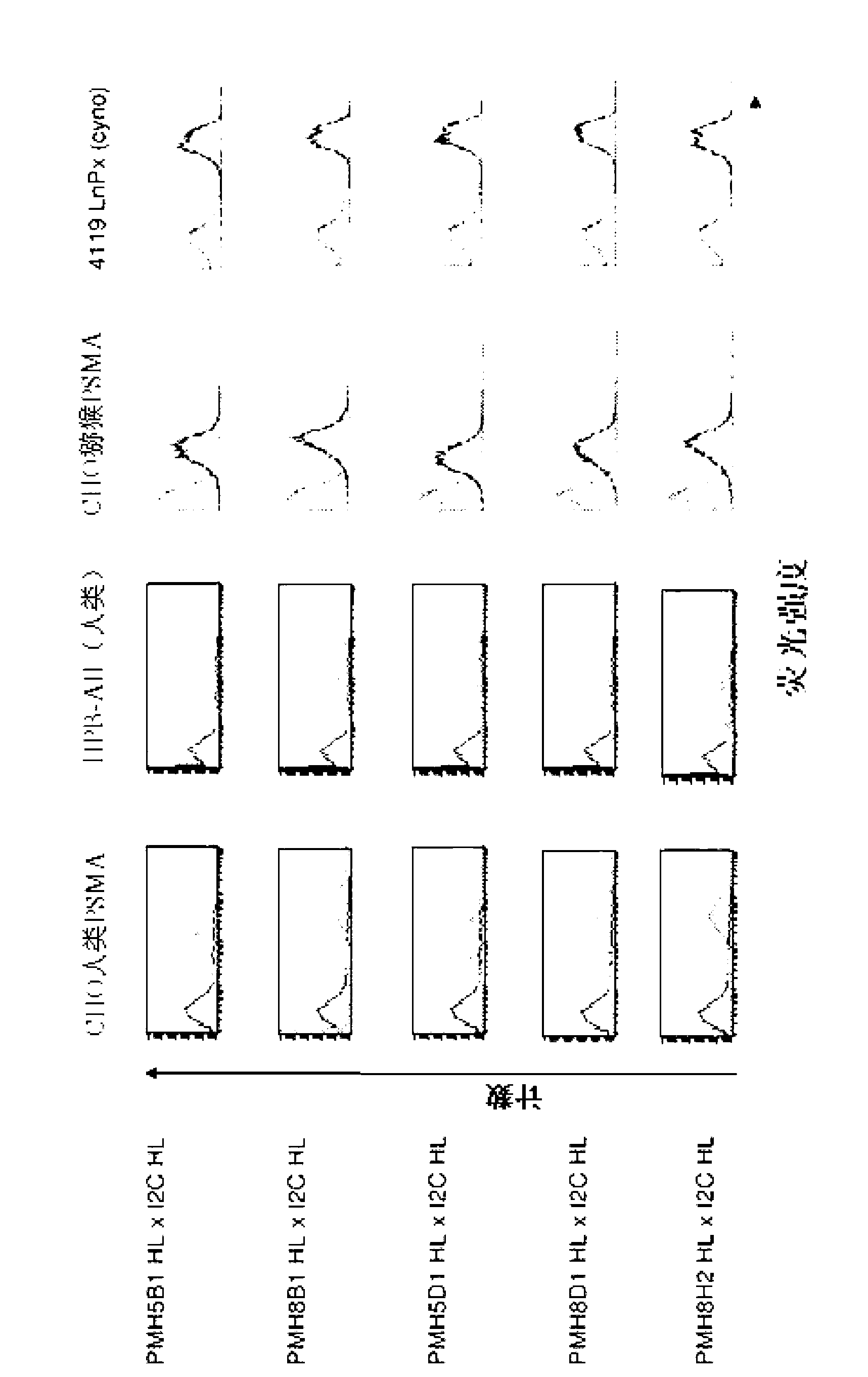
[0314] 2.1 Flow cytometry binding analysis of PSMA and CD3 cross-species specific bispecific antibody
[0315] To test the functionality of the cross-species specific bispecific antibody constructs with respect to binding ability to human and macaque PSMA and to human and macaque CD3, FACS analysis was performed. For this, CHO cells transfected with human PSMA and the human CD3-positive T-cell leukemia cell line HPB-ALL (DSMZ, Braunschweig, ACC483) were used to examine binding to human antigens. Generated by using macaque PSMA transfectants and macaque T cell line 4119LnPx (kindly provided by Prof. Fickenscher (Hygiene Institute, Virology, Erlangen-Nuernbe); published in Knappe A et al., and Fickenscher H., Blood 2000, 95, 3256 -61] to test the binding reactivity to macaque antigens.
[0316] Flow cytometry analysis was performed as follows:
[0317] Incubation of 200.000 cells of different cell lines with 50 μl of purified protein (2 μg / ml) of the cross-species specific bis...
Embodiment 3
[0326] Example 3 Binding assays of scFv against various cell lines
[0327] 3.1. Expression of single-chain antibody constructs in E. coli
[0328]The scFv molecules EpCAM 4-7 (WO 99 / 25818), PM74-G3, PM52-H3, PM52-C3, PM75-A10 and PM91-B6 are expressed by using the plasmid pComb3H5BFlag / His, where the expression construct (eg scFv) includes Flag -tag (DYKDDDDK) and His6-tag. Plasmid DNA of each scFv molecule was transformed into 100 [mu]l of heat shock competent E. coli TG1 and plated on carbenicillin LB-agar. E. coli transformed with pComb3H5BFlag / His containing VL- and VH-segments produced sufficient quantities of soluble scFv after induction with 1 mM IPTG. Due to an appropriate signal sequence, the scFv-chain is exported into the periplasm where it folds into a functional conformation.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com