Primer pair and standard substance for detecting mycobacteria and application thereof
A technology of mycobacteria and primer pairs, applied in the direction of microorganism-based methods, biochemical equipment and methods, microorganisms, etc., can solve the problems of gas phase mass spectrometry and liquid phase mass spectrometry, which are expensive and not suitable for routine detection, and achieve detection results Accurate, rapid detection, good specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0031] Embodiment 1, the preparation of specific primer pair and standard plasmid
[0032] 1. Preparation of standard plasmids
[0033] 1. Analyze the hsp65 gene sequences of all mycobacteria in NCBI, and obtain the DNA fragment shown in sequence 1 of the sequence table based on the analysis results. Sequence 1 has more than 98% homology with the hsp65 genes of various mycobacteria in NCBI, and has no homology with other genes. See Table 1 for partial homology comparison results.
[0034] Table 1 Homology comparison results between sequence 1 and some mycobacterial hsp65 genes
[0035]
[0036] 2. Synthesize the double-stranded DNA molecule shown in sequence 1 of the sequence listing.
[0037] 3. Insert the double-stranded DNA molecule synthesized in step 2 18-T Vector (TaKaRa Code: D101A) to obtain the recombinant plasmid pMD-18T-hsp65 (standard plasmid).
[0038] 2. Preparation of specific primer pairs
[0039] A pair of specific primers were designed based on seque...
Embodiment 2
[0043] Embodiment 2, establishment of mycobacterium quantitative PCR detection method
[0044] Mycobacterium fortuitum subsp.fortuitum: CGMCC number 1.513.
[0045] 1. Optimization of annealing temperature
[0046] 1. Extract the genomic DNA of Mycobacterium fortuitum.
[0047] 2. Using the genomic DNA extracted in step 1 as a template, using a primer pair composed of hsp65-F and hsp65-R, using Premix ExTaq TM (Code:DRR041S) and perform quantitative qPCR according to the instructions.
[0048] qPCR reaction system (20 μL): Premix ExTaq TM 10 μL, hsp65-F, hsp65-R, genomic DNA, make up the volume to 20 μL with double distilled water. The initial concentrations of hsp65-F and hsp65-R in the reaction system were both 0.2μmol / L.
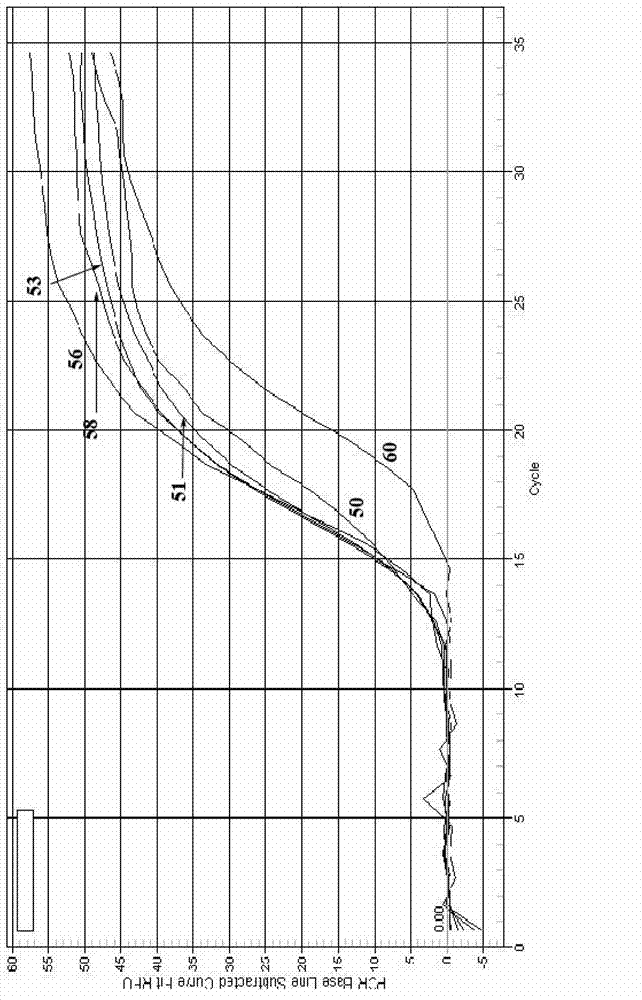
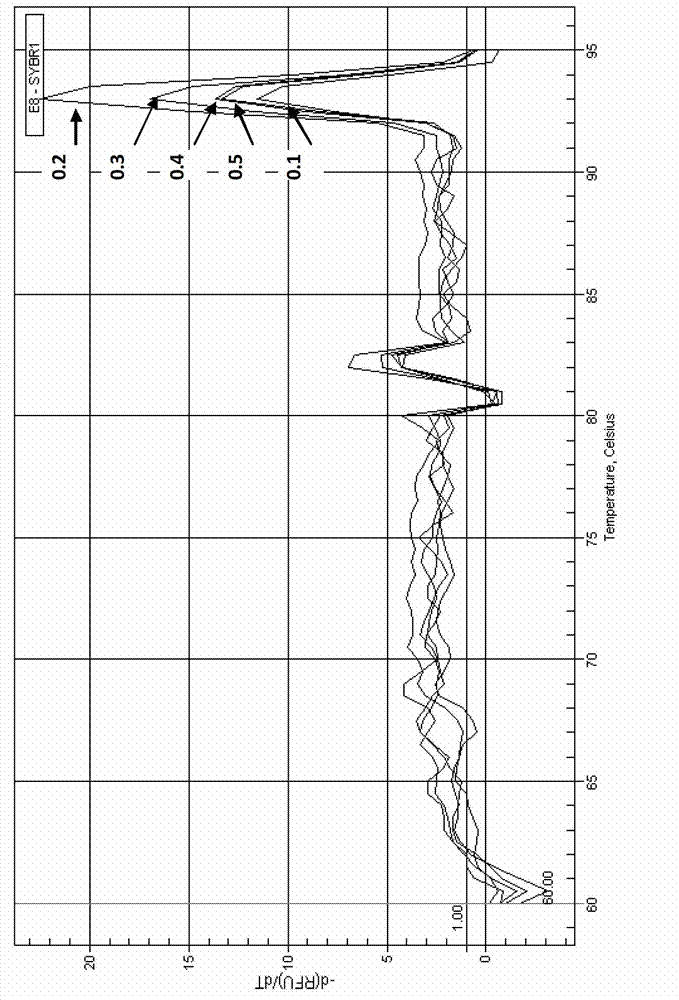
[0049] The reaction program of qPCR: (95°C, 5min) × 1 cycle; (95°C, 5s, annealing 30s, 72°C, 30s) × 40 cycles; fluorescence is collected during annealing; the process of melting curve is : 95°C, 1min, 65°C, 1min, starting from 65°C, the temperat...
Embodiment 3
[0057] Embodiment 3, the specificity experiment of specific primer pair
[0058] 1. Experimental samples
[0059] The experimental samples are as follows:
[0060] Escherichia coli (Escherichia coli): CGMCC number is 1.1369.
[0061] Salmonella typhimurium (Salmonella typhimurium): CGMCC number is 1.1194.
[0062] Enterococcus faecalis: CGMCC number is 1.2135.
[0063] Mycobacterium fortuitum subsp.fortuitum: CGMCC number 1.513.
[0064] Mycobacterium smegmatis (Mycobacterium smegmatis): CGMCC number is 1.2621.
[0065] Mycobacterium phlei: CGMCC number is 4.1180.
[0066] Mycobacterium diernhoferi: CGMCC number is 4.1179.
[0067] Mycobacterium vaccae: CGMCC number 4.1181.
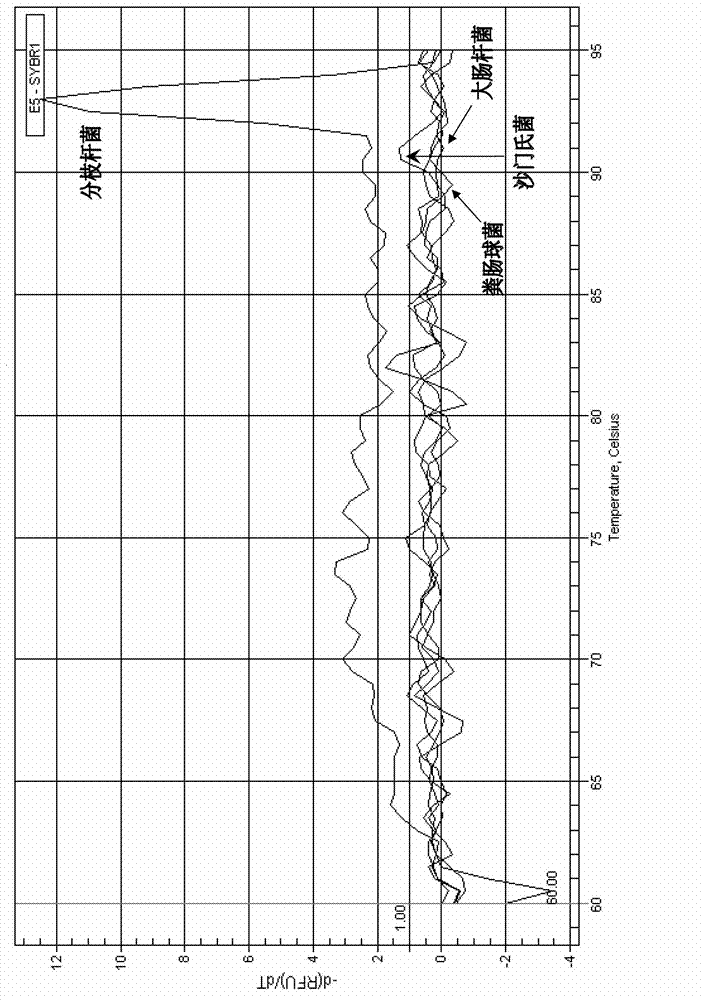
[0068] 2. Specificity experiment
[0069] 1. Extract the genomic DNA of each experimental sample.
[0070] 2. Using the genomic DNA extracted in step 1 as a template, using a primer pair composed of hsp65-F and hsp65-R, using Premix ExTaq TM Quantitative qPCR was performed according to the ins...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com