Defining diagnostic and therapeutic targets of conserved free floating fetal DNA in maternal circulating blood
A fetal and maternal technology, applied in the direction of organic compound library, biochemical equipment and methods, microbial determination/testing, etc., can solve problems such as the development of difficult sequences and mutation-specific assays
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0135] Example 1: Dx cleavage for fetal DNA extraction
[0136]Isolation of cell-floating fetal DNA from whole blood presents unique challenges. Two confounding variables that maximize fetal DNA yield from whole blood are the selective lysis and depolymerization of target-specific cells and DNA to efficiently extract them in the context of maternal genomic DNA. To accomplish this task, plan buffers and protocols that accomplish two key purposes. First, the gentle lysis protocol selectively lyses cells that are not in their optimal growth environment (i.e., the fetal trophoblast), thereby allowing the release of nucleic acids from the cells that are not otherwise present in the acellular DNA fraction, and second, depolymerization in A small DNA molecule that is not available for efficient extraction in its normal state. This lysis buffer and protocol increases the yield of fetal DNA in any given maternal whole blood sample by approximately 15%. After lysis, an automated met...
Embodiment 2
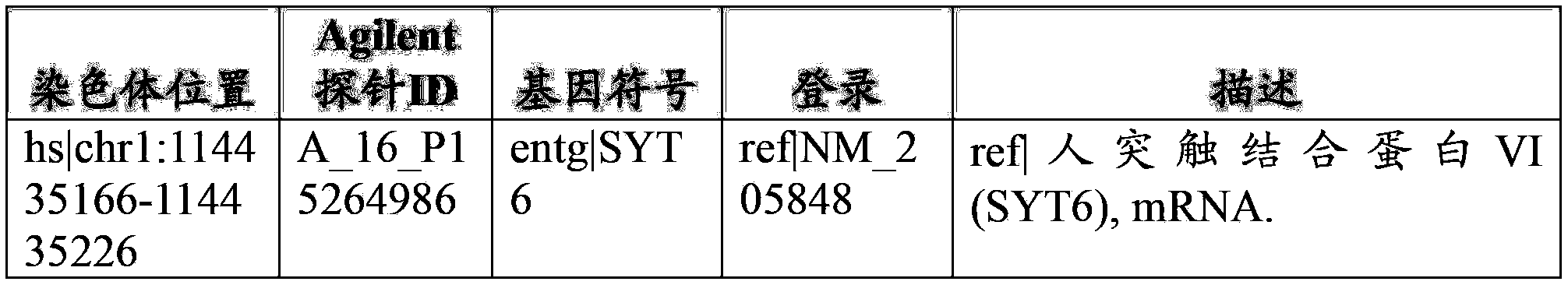
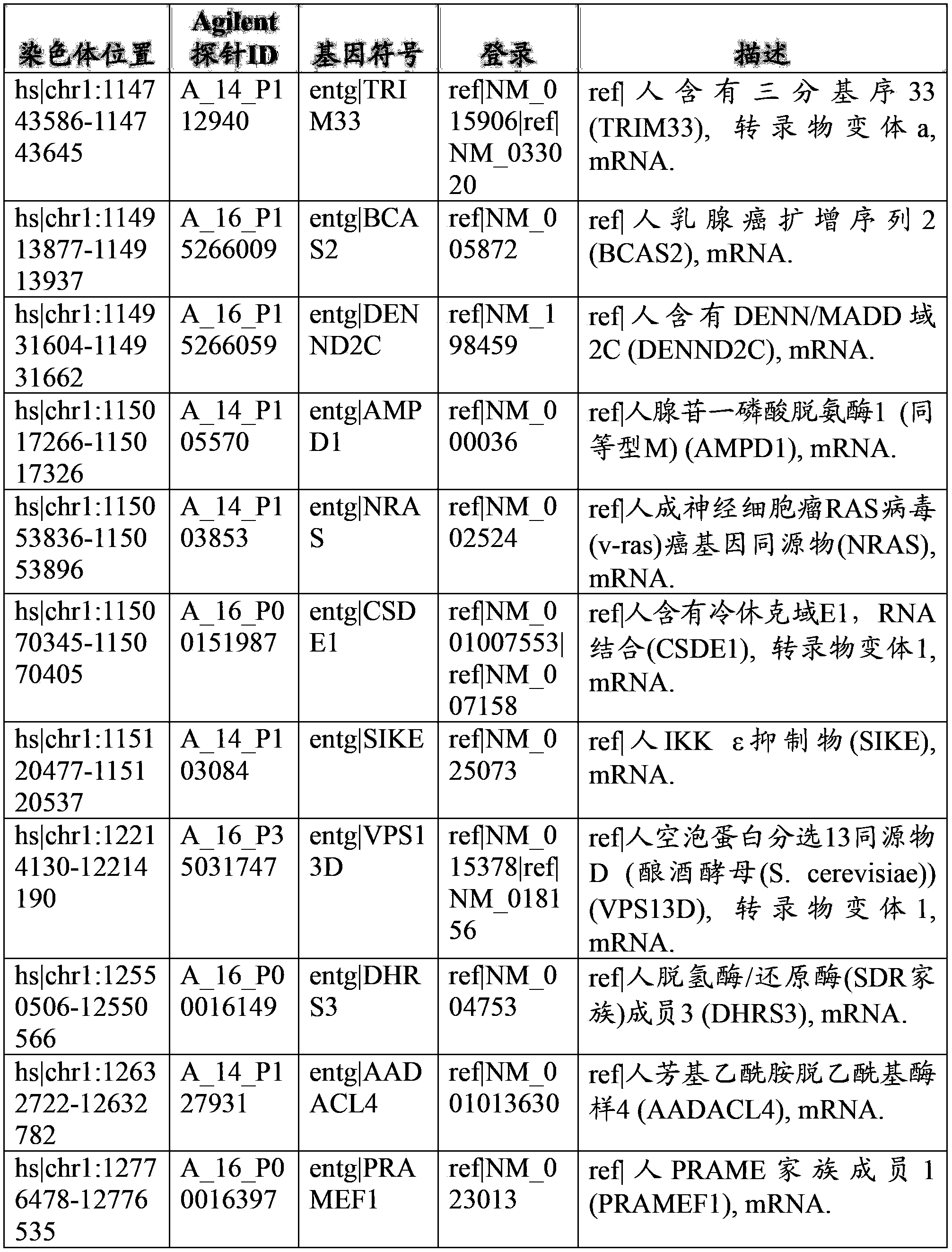
[0138] Example 2: Characterization of conserved episomal floating DNA sequences
[0139] Subtractive hybridization was used to identify fetal-specific sequences in Dx-cleaved, size-fractionated, cell-floating DNA. Briefly, this subtractive hybridization approach requires running two CGH arrays for each clinical situation. The first array analyzes maternal DNA against fetal DNA (products of conception), thereby identifying differences in fetal genomic DNA. A second array analyzes maternal DNA against enriched, free-floating fetal DNA (a product of maternal whole blood), thereby identifying regions present in free-floating fetal DNA. Comparative analysis of unique fetal segments from these two arrays identified conserved regions in the free-floating fetal DNA samples in each case analyzed. By following this hybridization protocol, we can confirm which sequences are present in the free-floating fetal DNA fraction when compared to the complete fetal genome. This is the first ...
Embodiment 3
[0144] Example 3: NextGen Sequencing
[0145] To fully understand the length and fidelity of sequences identified by array CGH, this NextGeneration sequencing approach was employed to validate and ultimately map conserved loci in the episomal floating fetal genome. The sequenced loci were derived from the conserved probe sequences identified using the array CGH described above. Briefly, conserved probe sequences identified as present in free-floating fetal DNA were used as "baits" to create capture libraries that were used to sequence entire segments of conserved free-floating fetal DNA. The degree of natural genomic variation among individuals creates additional problems in predicting the conservation of fetal DNA among individuals. Therefore, it is prudent to use constitutive ("normal") DNA from the same individual as well as fetal DNA as a potential reference, in this case, it is maternal DNA. For DNA analysis, using paired-end genomic libraries, a targeted sequencing a...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com