Detection reagent for Shigella flexneri plasmid-carried gene Ipt-O and application thereof
The technology of Shigella flexneri and detection reagent is applied in the field of identification of Shigella flexneri MASF IV-1 phenotype and detection primers for Shigella flexneri plasmid carrying gene lpt-O, which can solve the problem of antigenic determination Factors are not found and other problems, to achieve the effect of simple judgment, avoiding subjectivity, and accurate identification
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0060] Embodiment 1, the design of lpt-O gene-specific detection primer
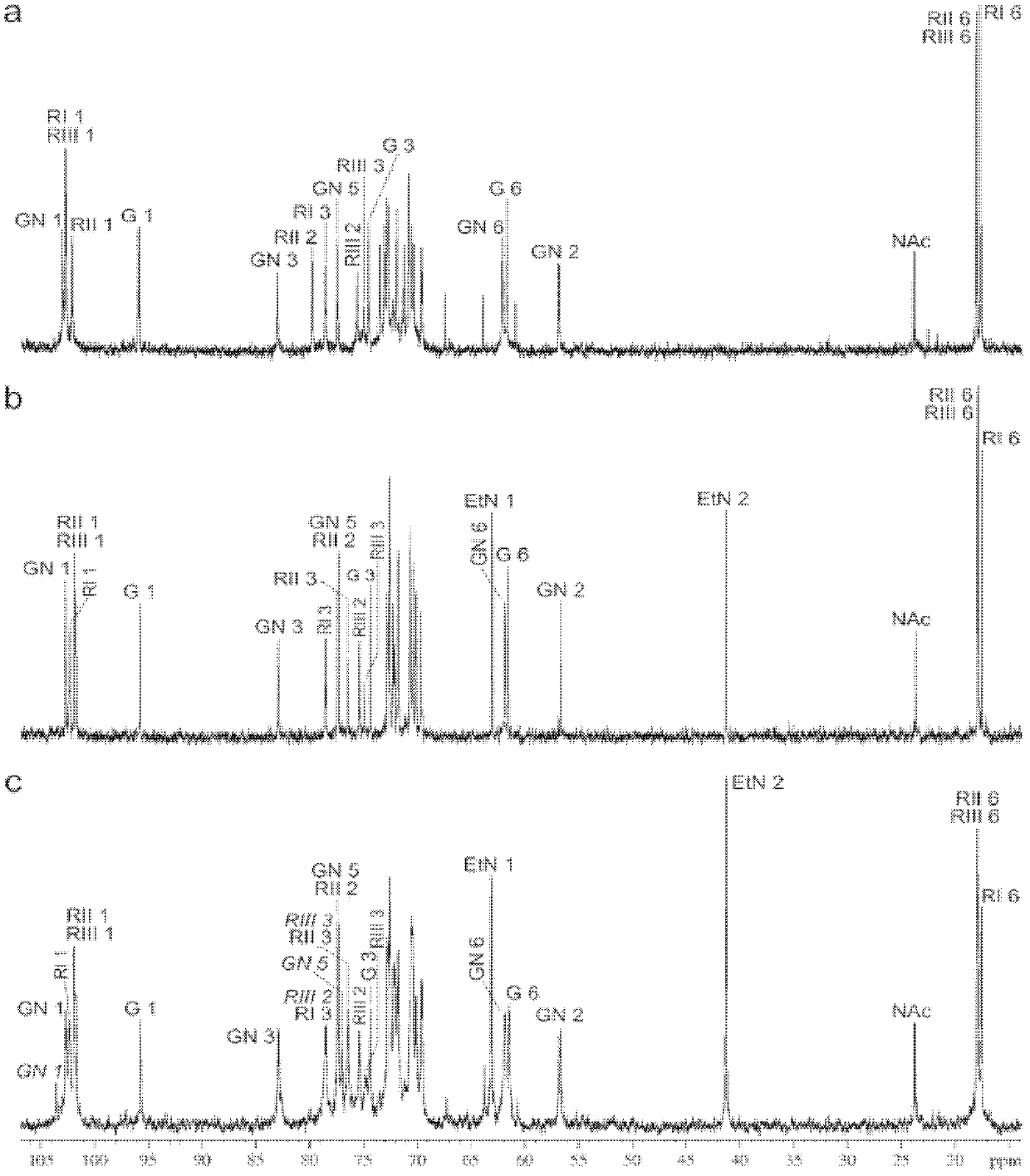
[0061] In this embodiment, a two-dimensional (two-dimensional) 1 H, 1 H and 1 H, 13 C NMR technology (Duus et al., 2000), analyzed the LPS structure of Xv serotype strain 2002017, and its 1 H NMR and 13 C NMR spectrum see figure 1 In b, it has a typical X serotype O antibody structure (Kenne et al., 1977) ( figure 2 Middle B), in addition, in the NMR spectrum of Xv bacterial strain, find the signal of a PEtN group (Table 1), this PEtN group adds in RhaII ( figure 2 Middle B), and this is the same as serotype X ( figure 2 The only difference in A). Considering the serotype difference between the two, it can be judged that PEtN modification is the reason for the appearance of MASF IV-1+ phenotype.
[0062] Table 1 1 H and 13 C NMR chemical shift s(δ, ppm)
[0063]
[0064] In the present invention, a gene SFxv_5135 and PEtN transferase protein Lpt3(N.meningitides), LptA(N.meningitides),...
Embodiment 2
[0071] Embodiment 2, PCR amplification detection distinguishes Xv and X serotype bacterial strain
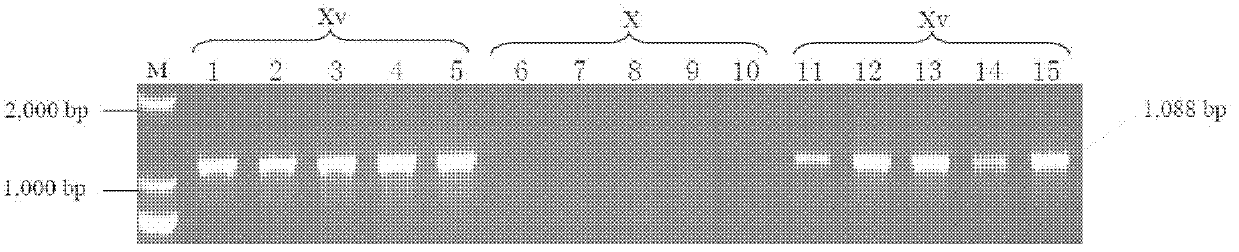
[0072] In this example, 10 strains of Shigella flexneri serotype Xv and 5 strains of X serotype were detected by using the designed primer pair lpt-O-2.
[0073] PCR amplification:
[0074] The primer pair was commissioned to Sangon Biotech (Shanghai) for synthesis. PCR amplification was carried out using a PCR amplification kit from TaKaRa Company (KakaRa, Japan). Each PCR reaction mixture contains: 1×PCR buffer, 0.2 μM of primers, 3 μl of template DNA, 2.5U DNA polymerase and 0.4 mM dNTP, total 50 μl. PCR amplification was carried out using PCR instrument SensoQuest LabCycler (Germany), and the specific conditions were: pre-denaturation at 94°C for 5 min; denaturation at 94°C for 30 s, annealing at 55°C for 50 s, and extension at 72°C for 2 min, a total of 30 cycles; finally, extension at 72°C for 5 min.
[0075] The amplified products were subjected to electrophoresis, and...
Embodiment 3
[0076] Embodiment 3, PCR amplification detects MASF IV-1 phenotype
[0077] In the present embodiment, according to the amplification method described in Example 2, the 333 strains listed in Table 2 include 15 serotype strains for PCR amplification detection, and the results are only positive in MASF IV-1 strains (26Xv, 1 strain 4av and 25 strains Yv) and 4 strains X, 1 strain 4b, confirmed by sequencing, a single base deletion mutation occurred in the lpt-O gene in these 4 strains X and 1 strain 4b (Table 2) ( Figure 7 , Figure 8 ).
[0078] Carrying situation of table 2 lpt-O gene in Shigella flexneri
[0079]
[0080]
[0081] 1 + and - / + indicate the presence of the lpt-O gene. The number of positive strains is shown in parentheses.
[0082] 2 4av serotype strains (2002091 and NCTC 8296) and 4a serotype strains were differentially agglutinated in MASF IV-1.
[0083] 3 The serotype difference between Yv serotype strains and Y serotype strains lies in MASF IV-1 ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| separation | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com