RT-PCR method integrated with restriction endonuclease removal of DNA pollution
A technology of RT-PCR and restriction endonuclease, applied in the field of RT-PCR that integrates restriction endonuclease to remove DNA contamination, can solve the problems of general limitation, application limitation, cost increase, etc., and achieve PCR amplification efficiency Unaffected, avoiding cross-contamination interference, and reducing the effect of cross-contamination of PCR products
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
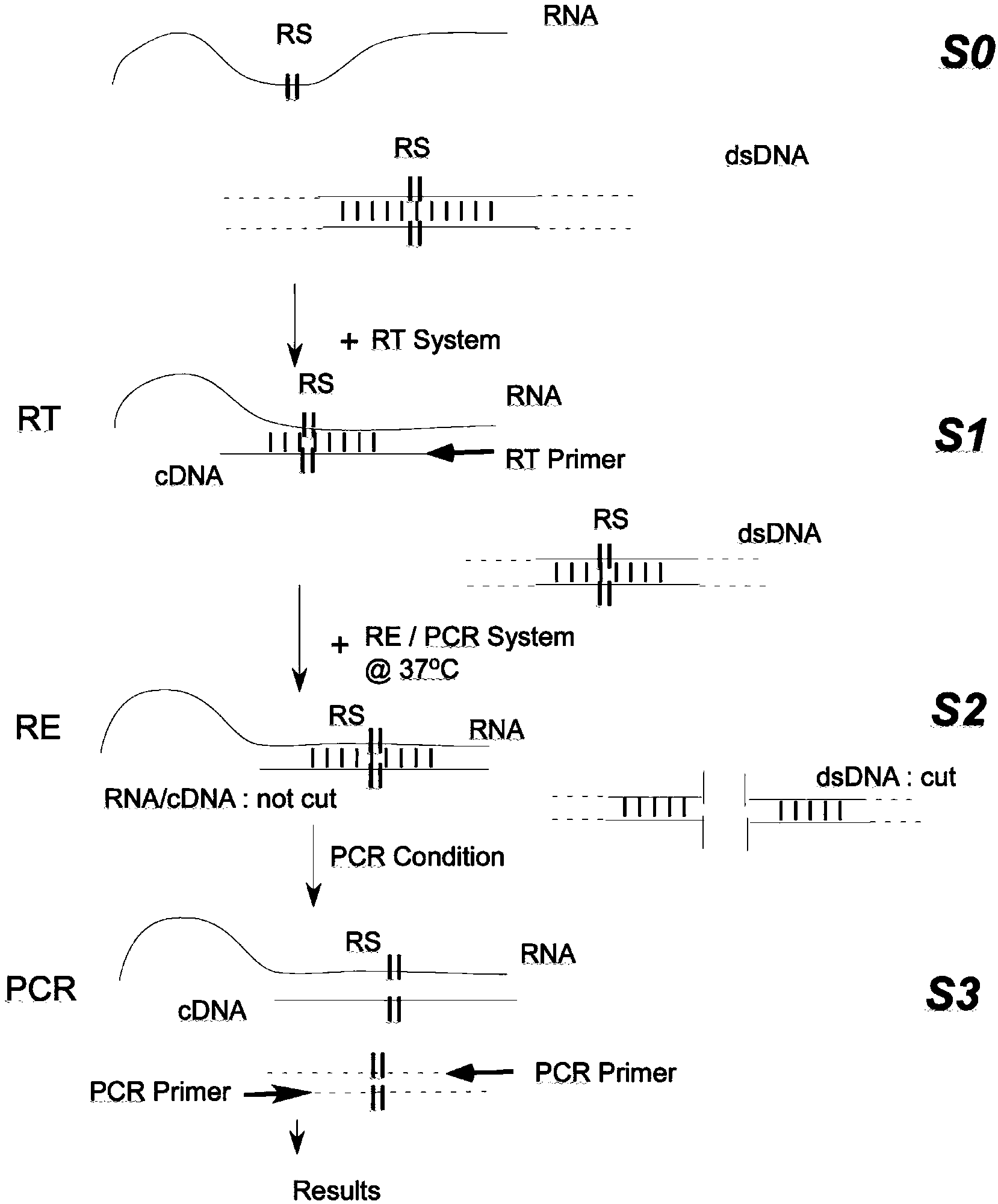
[0040] Example 1: The RDIRD system integrated with BamHI can remove the interference of trace genomic DNA in RNA samples on hTERT mRNA RT-PCR
[0041] The region between 2525-2615 on hTERT mRNA (NCBI NM_001193376) was selected as the RT-PCR amplification detection region, which contained a BamHI site (2547-2552) and a PstI site (2581-2586). RT and PCR primer sequences are described in the Summary of the Invention section.
[0042] Preparation of RNA samples: human skin cancer cell line A549 cells were cultured in a 24-well plate, 10,000 cells / well, after overnight culture, the culture medium was aspirated, and 200ul Trizol lysate (Invitrogen Company) was added, and pipetting was repeated. Transfer to a 1.5ml centrifuge tube, vibrate vigorously on a vortex shaker at room temperature for 10min, after a short centrifugation, add 50ul chloroform, vibrate vigorously for 1min, let stand at room temperature for 2min, centrifuge at 4°C, 15000rpm for 20min, take the upper aqueous phase...
Embodiment 2
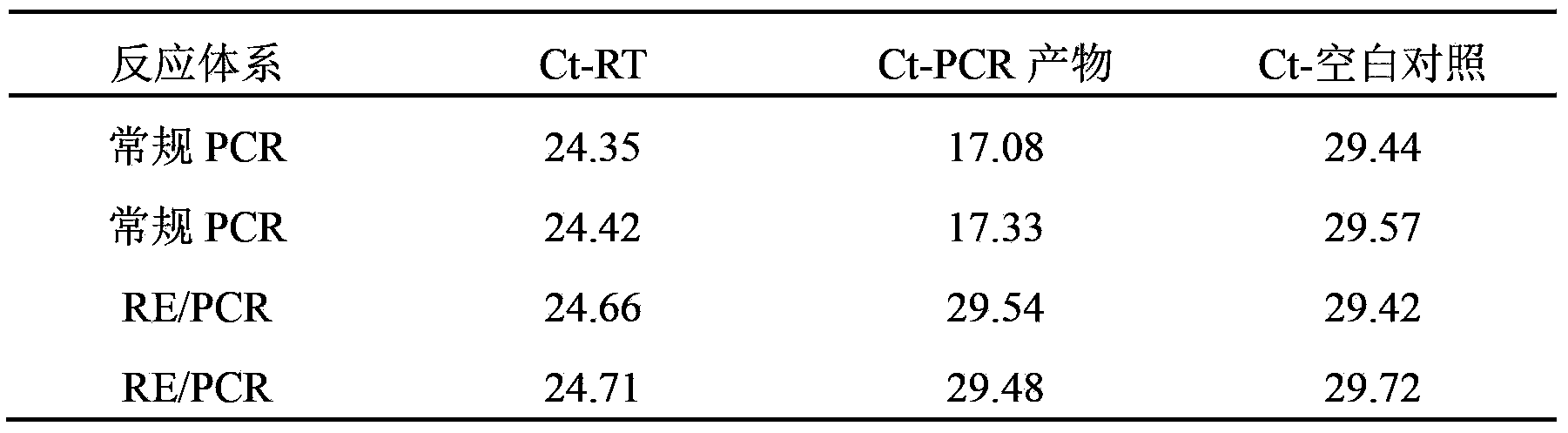
[0048] Example 2: The RDIRD system integrated with BamHI can remove the interference of trace PCR product contamination on hTERTmRNA RT-PCR
[0049] Amplified region, primers, preparation of RNA sample, RT reaction, etc., are the same as in Example 1, PCR without RT group, the PCR product diluted 10,000 times (obtained from Example 1) was used as a template instead of the RNA sample, and the results are as follows:
[0050]
[0051] It can be seen that the Ct value of the blank control (derived from the primer dimer) is between 29.72 and 29.42. Using conventional PCR, the Ct value of the 10,000-fold diluted PCR product (Ct-PCR product) is between 17.33-17.08, which is significantly smaller than the Ct value of the blank control, indicating that it is sufficient if there is a trace of PCR product mixed into the conventional PCR system Significant amplification was formed; while the value of the Ct-PCR product obtained by the RE / PCR system integrated with BamHI was between 29...
Embodiment 3
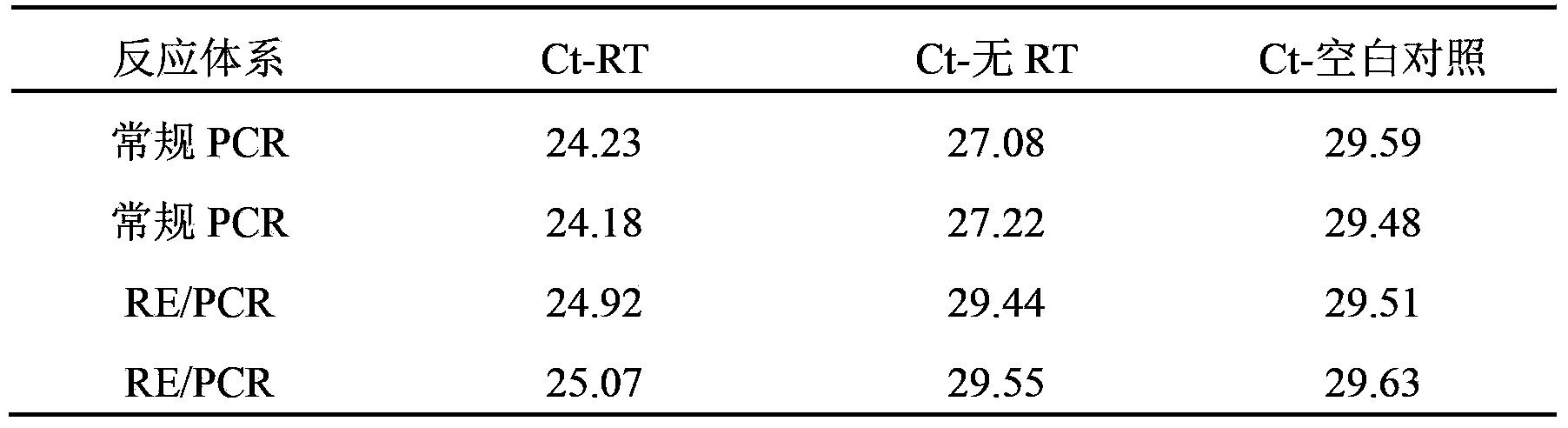
[0052] Example 3: The RDIRD system integrated with PstI can remove the interference of trace genomic DNA in RNA samples to hTERT mRNA RT-PCR
[0053] Amplified regions, primers, preparation of RNA samples, RT reaction, PCR, etc., are the same as in Example 1, except that BamHI is replaced by PstI in the RE / PCR system of RDIRD, and the results are as follows:
[0054]
[0055] It can be seen that the Ct value of the blank control (derived from the primer dimer) is between 29.65 and 29.47. Using conventional PCR, the Ct value of the RNA sample without RT reaction (Ct-no RT) is between 27.26-27.16, which is significantly smaller than the Ct value of the blank control, indicating that there is a trace amount of genomic DNA mixed in the RNA sample to be amplified and the value of Ct-no RT obtained by the RE / PCR system integrated with PstI is between 29.52 and 29.39, which is equivalent to the Ct value of the blank control, indicating that this system can effectively cut a trace ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com