Specific identification for transgenic rice kefeng No.6 strain via adoption of RPA (Recombinase Ploymerase Amplification) technology
A technical identification and transgenic technology, which is applied in the field of primer and probe combination, can solve the problems that cannot be further satisfied with the rapid detection of transgenic products, and achieve the effect of high sensitivity and improved ability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
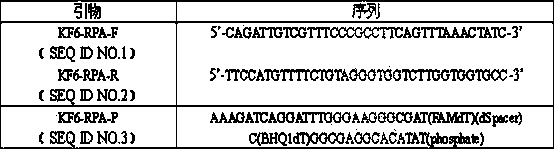
[0017] Example 1: Design and screening of primers
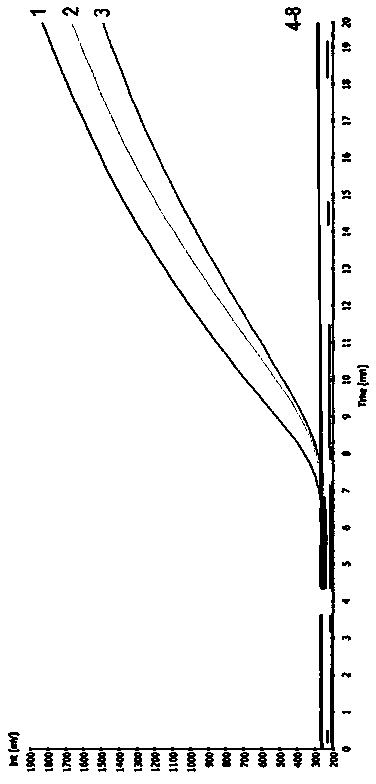
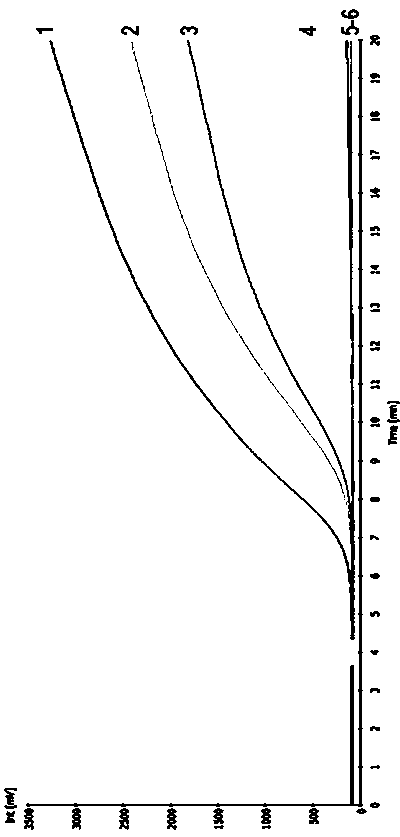
[0018] First of all, primer design and screening: RPA primer design usually needs to consider the following factors: (1) GC content is 40%-60%; (2) Try to avoid secondary structures inside the primer; (3) Avoid primer duplication sequence. RPA requires a primer length of 30-35nt. In the experiment, multiple pairs of primers need to be designed from both ends of the target sequence for optimization, screening, and the replacement or increase or decrease of individual bases will have an important impact on the experimental results. In this experiment, a probe was designed according to the specific sequence of Kefeng 6 transformant (GenBank No. HM124448), and 4 upstream primers and 5 downstream primers were designed on both sides of the probe. Using 500 copies of Kefeng 6 genomic DNA as a template, different primer combinations were screened for fluorescence, and finally a pair of primers with short amplification take-off tim...
Embodiment 2
[0022] Example 2: Detection of Kefeng 6 with designed primers
[0023] 1. Experimental Materials
[0024] (1) Plant material
[0025] Genetically modified rice Kefeng No. 6 (leaf), genetically modified rice Kefeng No. 6 seed powder (content 5%), genetically modified rice Kefeng No. 6 seed powder (content 1%), genetically modified rice Kefeng 1 (leaf), genetically modified rice TT51-1 (leaf), transgenic rice Kangyou 97 (leaf), non-transgenic rice Minghui 63 (leaf).
[0026] (2) Enzymes and reagents
[0027] Molecular biology reagents, TwistAmp DNA Amplification Exo Kits were purchased from TwistDX Company, and other biochemical reagents were imported or domestic analytically pure. Primers were synthesized by Beijing Sangon Biotechnology Co., Ltd.
[0028] (3) Experimental equipment
[0029] DNA processing instrument: low temperature mixing ball mill MM400 (Retsch)
[0030] Fluorescence detector: RPA amplification detector (Twista) or fluorescent quantitative PCR instru...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com