Gibson assembly carrier, preparation method therefor and applications thereof
A carrier and application technology, applied in the field of DNA synthesis, can solve the problems of cumbersome cloning process and instability, and achieve the effect of simple design and efficient assembly results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
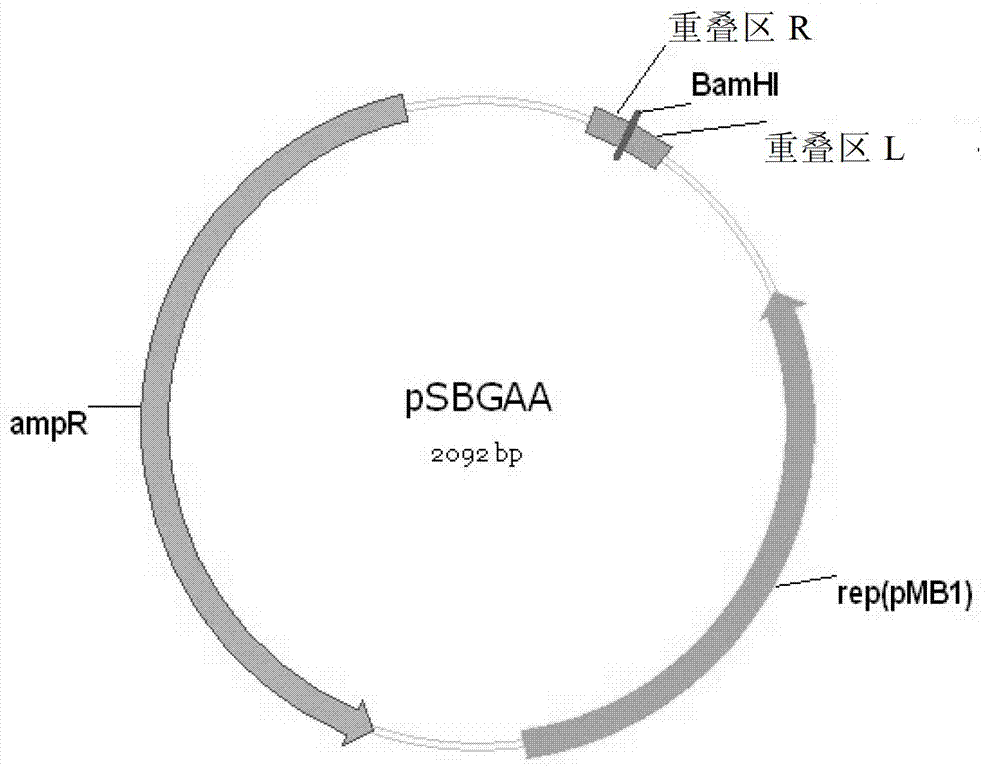
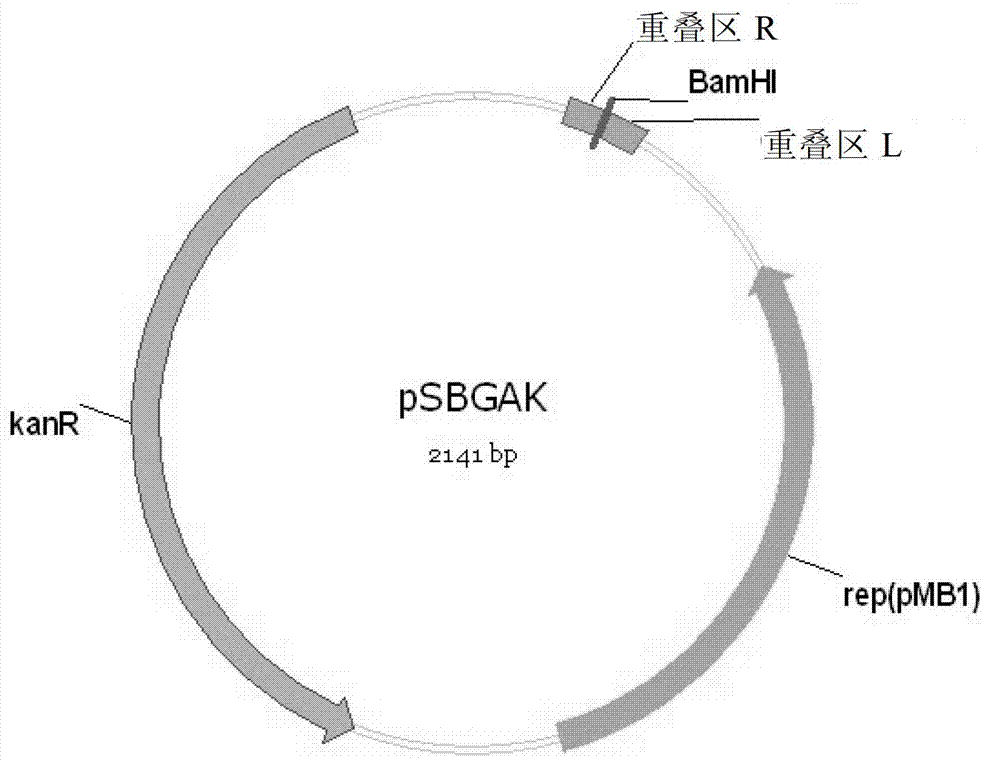
[0056] Example 1 - Preparation of vectors pSBGAA and pSBGAK
[0057] The specific implementation is as follows:
[0058] 1) Use the vectors pSB1A3 and pSB1K3 as templates for PCR amplification:
[0059] The reaction system includes: Ex Taq (5U / μL) 0.25μL, 10×PCR buffer 2.5μL, primers pSB-F, pSB-R 1μL each, dNTP (each 2.5mM) 2μL, DNA template 1μL, ddH 2 O17.25 μL.
[0060] Reaction program: pre-denaturation at 94°C for 4 min; denaturation at 94°C for 30 sec, annealing at 55°C for 30 sec, extension at 72°C for 2 min, 30 cycles; extension at 72°C for 5 min. The PCR product was recovered with a Qiagen PCR purification kit, and the method was referred to its instruction manual.
[0062] Reaction procedure: BamHI (15U / μL) 1 μL, 10×K buffer 2 μL, recovered PCR product DNA 1 μg, add ddH2O to make up 20 μL. Enzyme digestion at 30°C for 3h. The digested product was recovered using Qiagen PCR purification kit, and the method was referred to its instruct...
Embodiment 2
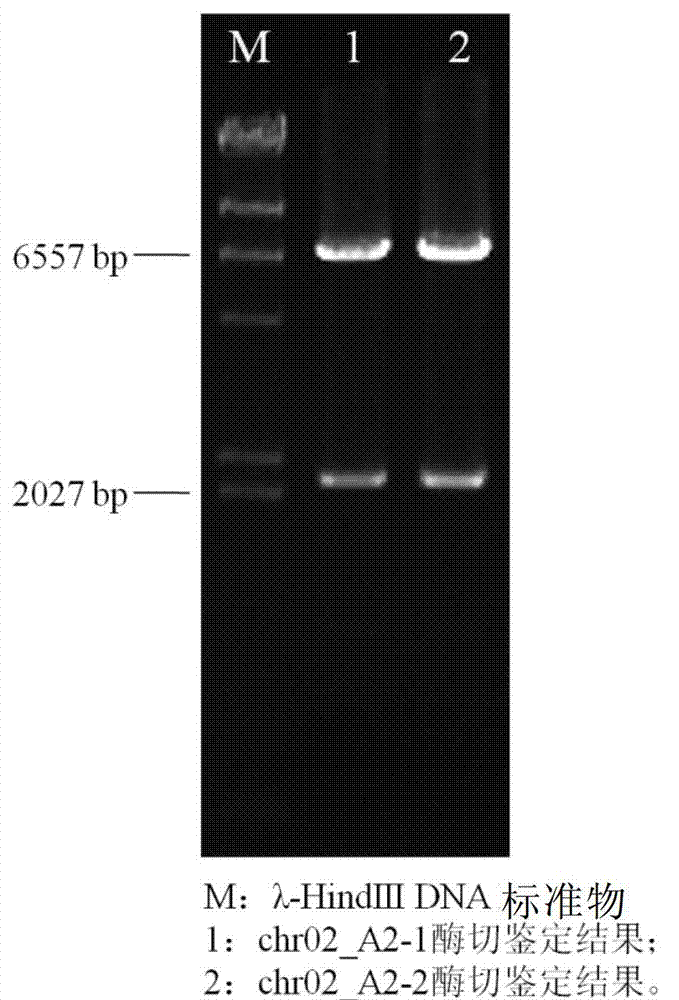
[0073] Example 2 - Assembly of long yeast DNA fragments using the vector pSBGAA
[0074] 1) Sequence design:
[0075]Select the DNA fragment of yeast chromosome 2, numbered yeast_chr02_3_22.A2, with a length of about 6.8kb, add the overlapping region R and overlapping region L of the vector to both ends of the DNA fragment, and then divide the DNA fragment into three fragments of about 2kb, named respectively They are yeast_chr02_3_22.A2.F1, yeast_chr02_3_22.A2.F2, yeast_chr02_3_22.A2.F3, with a 40bp homologous region in the middle. Add an XhoI restriction site at the end of the fragment and send it to Gene Synthesis Company for synthesis.
[0076] 2) Assembly of DNA fragments and vector preparation:
[0077] The synthesized DNA fragment and the constructed vector were enzymatically cut into linear DNA fragments respectively.
[0078] Vector digestion: BamHI (15U / μL) 1μL, 10×K buffer 2μL, pSBGAA 1μg, add ddH 2 Make up 20 μL with O; digest at 30°C for 3 hours.
[0079] Dig...
Embodiment 3
[0092] Example 3 - Assembly of long yeast DNA fragments using vector pSBGAK
[0093] 1) Sequence design:
[0094] Select the DNA fragment of yeast chromosome 2, numbered yeast_chr02_3_22.A5, with a length of about 11kb, and add the overlapping region R and overlapping region L of the vector to both ends of the DNA fragment, and then divide the DNA fragment into five fragments of about 2kb, named respectively Yeast_chr02_3_22.A5.F1, yeast_chr02_3_22.A5.F2, yeast_chr02_3_22.A5.F3, yeast_chr02_3_22.A5.F4, yeast_chr02_3_22.A5.F5 have a homologous region of 40 bp in the middle. Add an XhoI restriction site at the end of the fragment and send it to Gene Synthesis Company for synthesis.
[0095] 2) Assembly of DNA fragments and vector preparation:
[0096] The synthesized DNA fragment and the constructed vector were enzymatically cut into linear DNA fragments respectively.
[0097] Vector digestion: BamHI (15U / μL) 1μL, 10×K buffer 2μL, pSBGAK 1μg, add ddH 2 Make up 20 μL with O; ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com