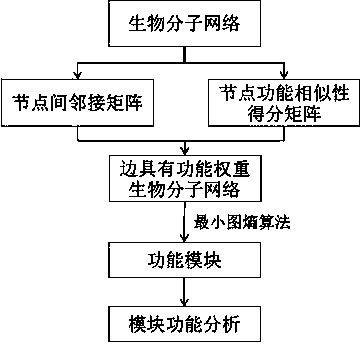
Biomolecular network analysis method based on function module
A biological network and functional module technology, applied in the field of biomolecular networks such as protein-protein interaction networks or gene expression regulatory networks, can solve the problem of not considering the functional similarity of adjacent nodes, and achieve the effect of high functional correlation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0043] Embodiment 1. Taking the gene and protein interaction network composed of a group of gene chip expression data after administration and treatment of nifedipine in the hypertensive mouse model as an example, the method of the present invention is specifically described:
[0044] Integrating protein interaction data in protein interaction databases such as BIOGRID, INTACT, MINT, and NIA Mouse Protein-Protein Interaction Database, after removing duplicate data and self-interactions, a global mouse gene and protein interaction network is obtained. Due to the limited data of this kind, the interaction data of orthologous and paralogous proteins of mouse proteins in other model organisms were used to predict the interaction of these proteins in mice, and a total of 65,850 small Mouse protein interaction data.
[0045] By performing mean centered normalization on the gene chip data, the genes whose normalized expression value was greater than 1 were regarded as expressed genes...
Embodiment 2
[0059] Embodiment 2. Based on the 39,240 protein interactions verified by experiments provided in the human Human Protein Reference Database (HPRD) database, 3000 protein interaction relationships were randomly screened, and protein self-interactions were removed to obtain 1,478 protein interactions from 2,095 edges. A network G of protein nodes.
[0060] Step 1: Calculate the adjacency matrix M of the network G adj ,M adj It is a matrix of 1,478 rows and 1,478 columns, each row and each column represent a unique gene, if there is an interaction edge between two proteins in the random network G, the corresponding element in the matrix is 1, otherwise it is 0.
[0061] Step 2: Using R's GOSemSim software package, based on the biological process in Gene Ontology (GO), calculate the semantic similarity score M between 1,478 proteins sim ,M sim The median value of 0.307.
[0062] Step 3, calculate the function weight matrix M of the edge of the network G E , , " "Repres...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com