Method and kit for characterizing rna in a composition
A composition and hybrid technology, applied in the field of expression profiling, can solve the problems of time-consuming, expensive, and error-prone comparative research in working steps
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0134] Example 1 - mRNA profile obtained by hybrid capture technology:
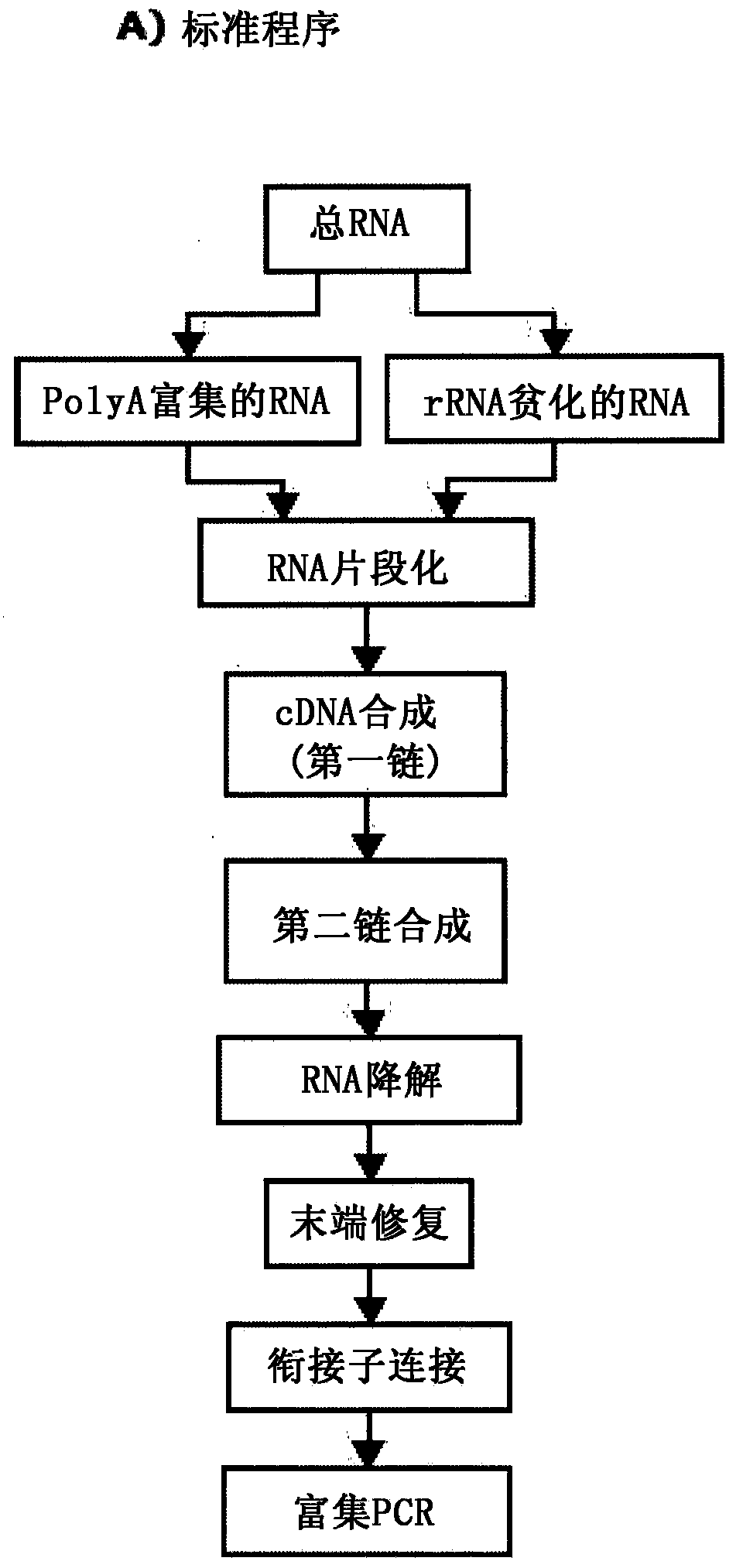
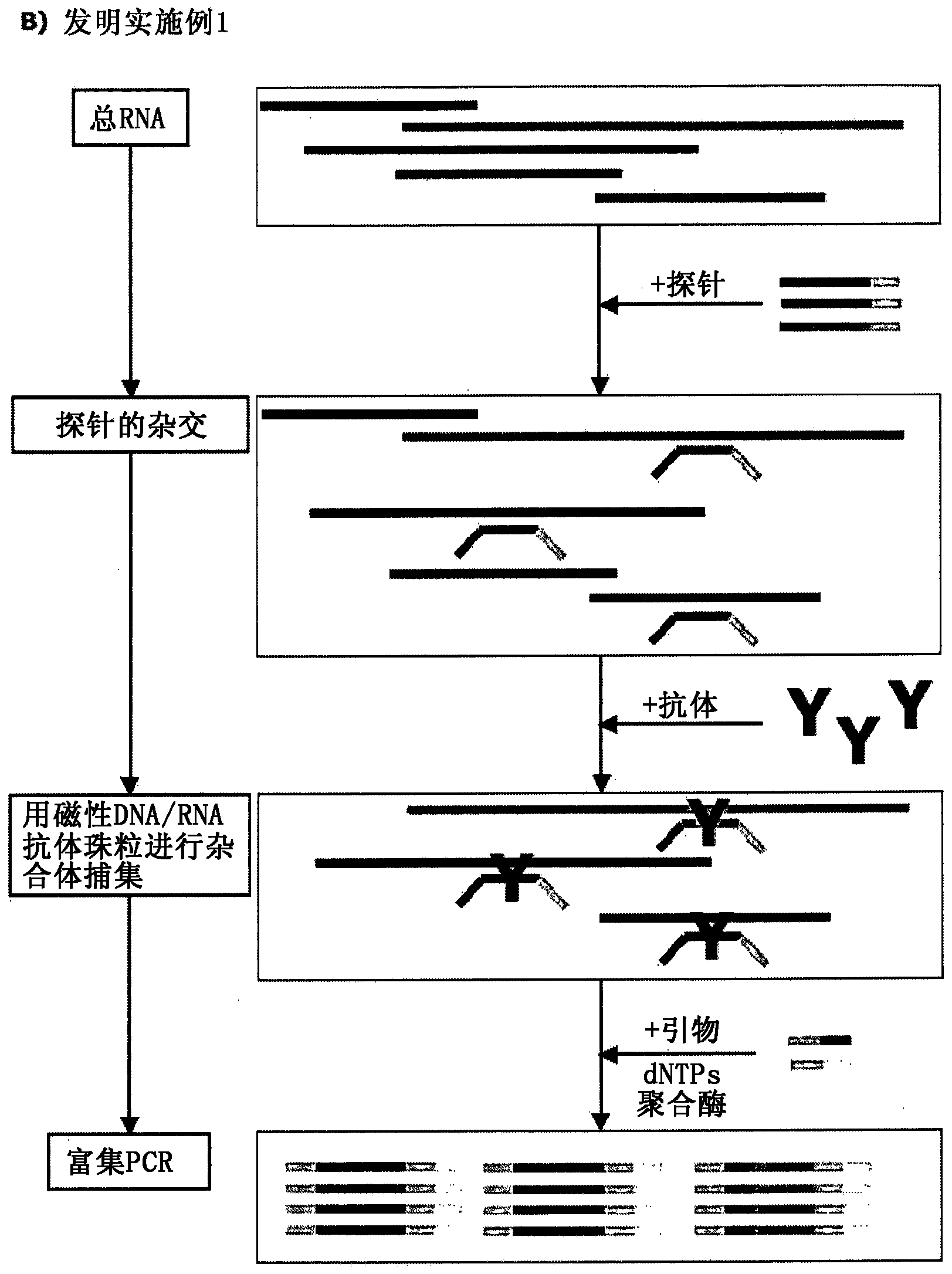
[0135] Specific DNA oligonucleotides containing universal adapters at the 5' and 3' ends are hybridized to the target mRNA and then captured with antibodies that bind the DNA / RNA hybrid. After magnetic isolation of DNA / RNA hybrid molecules, the purified probe library was enriched by PCR prior to sequencing ( figure 1 ).
[0136] DNA probes used to hybridize to target mRNAs are specifically designed to have comparable thermodynamic properties. Hybridization of RNA to excess oligonucleotides prior to purification of DNA / RNA hybrids allows quantification of target mRNA by determining the number of DNA probes via sequencing.
[0137] Selectivity for different mRNAs can be increased by placing probes at exon-exon junctions and adjusting appropriate hybridization conditions. In addition, it allows obtaining expression profiles of different splice variants of mRNA.
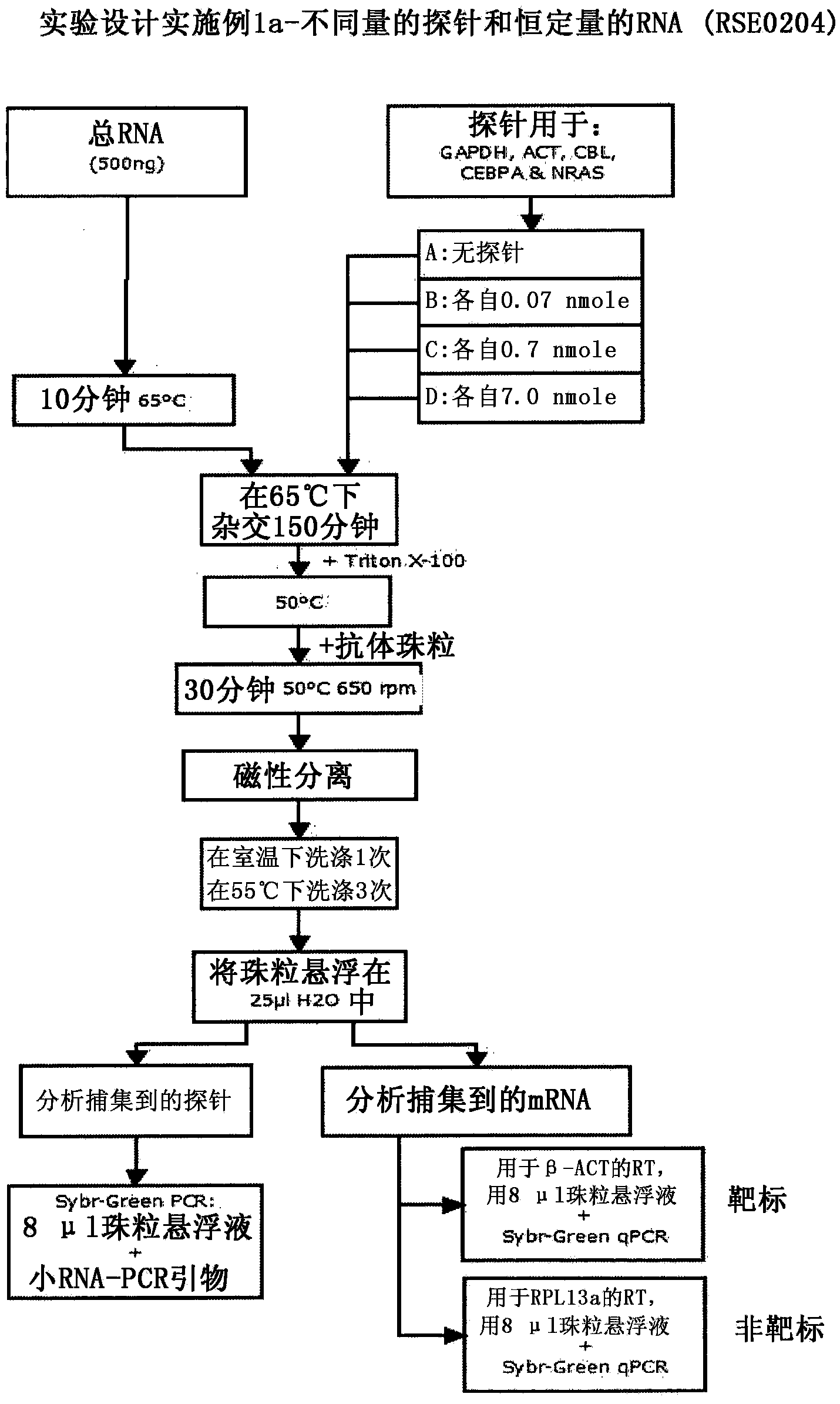
[0138] Example 1 - Experiment 1: Hybridi...
Embodiment 2
[0157] Example 2 - mRNA profiles obtained by ligation of oligonucleotide probes on RNA templates:
[0158] principle:
[0159] The probe consists of two tailed oligonucleotides. OligoB contains a universal 5' tail and a target-specific 3' sequence. OligoA consists of a target-specific 5' end and a general 3' tail. The two tails differ in their base composition. In addition, oligoA at the 5' end is phosphorylated. The two oligomers are matched in a gap-free, directly adjacent manner on their target RNA molecules, allowing the 3' end of oligoB to ligate to the phosphorylated 5' end of oligoA. After hybridization and ligation, the fusion oligo probes can be amplified via standard PCR using sequencer platform-specific enrichment primers ( Figure 9 ). Probes that do not hybridize in immediate proximity and / or in the correct order will not be amplified by enrichment PCR. Subsequently, the enriched probes can be sequenced. Probe design follows traditional primer design rules...
Embodiment 3
[0165] Example 3 - mRNA profiles obtained by hybridization, reverse transcriptase reaction and subsequent ligation of tailed oligonucleotide probes on RNA templates:
[0166] principle:
[0167] Probes were designed as in Example 2, but oligoA and oligoB were separated by a significant distance to match their RNA targets. Therefore, after hybridization, a polymerase step is required to shorten the gap between the two probe oligomers prior to ligation ( Figure 12 ).
[0168] Compared to the previous examples, this additional DNA synthesis step offers several advantages:
[0169] - Improved selectivity for generating probe fragments that can be amplified by PCR. Generation of PCR amplifiable fragments requires proper priming of the two probe oligomers, proper filling of the gap between the two oligomers and ligation of the target-specific oligomeric ends.
[0170] - The number of probes for the complete splicing profile of the target gene can be reduced, since one probe pai...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com