Double-labelling fluorescence activation type probe and method for detecting DNA by adopting same
A fluorescence-activated and labeled fluorescent technology, applied in the field of double-labeled fluorescent-activated probes, high-sensitivity analysis and detection of DNA, can solve the problems of detection sensitivity constraints, lack of signal amplification mechanism, etc., and achieve simple structure, simplified operation steps, and high sensitivity Detection effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
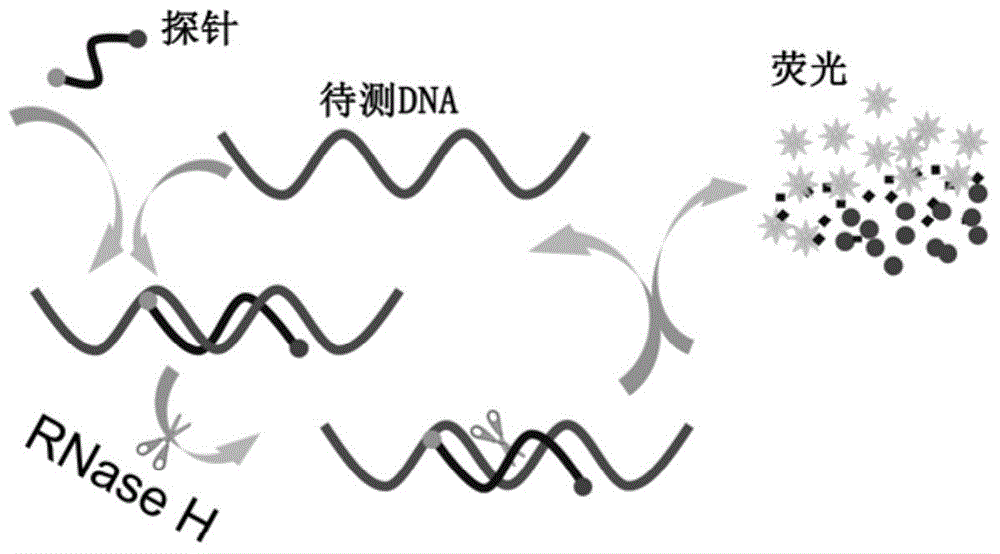
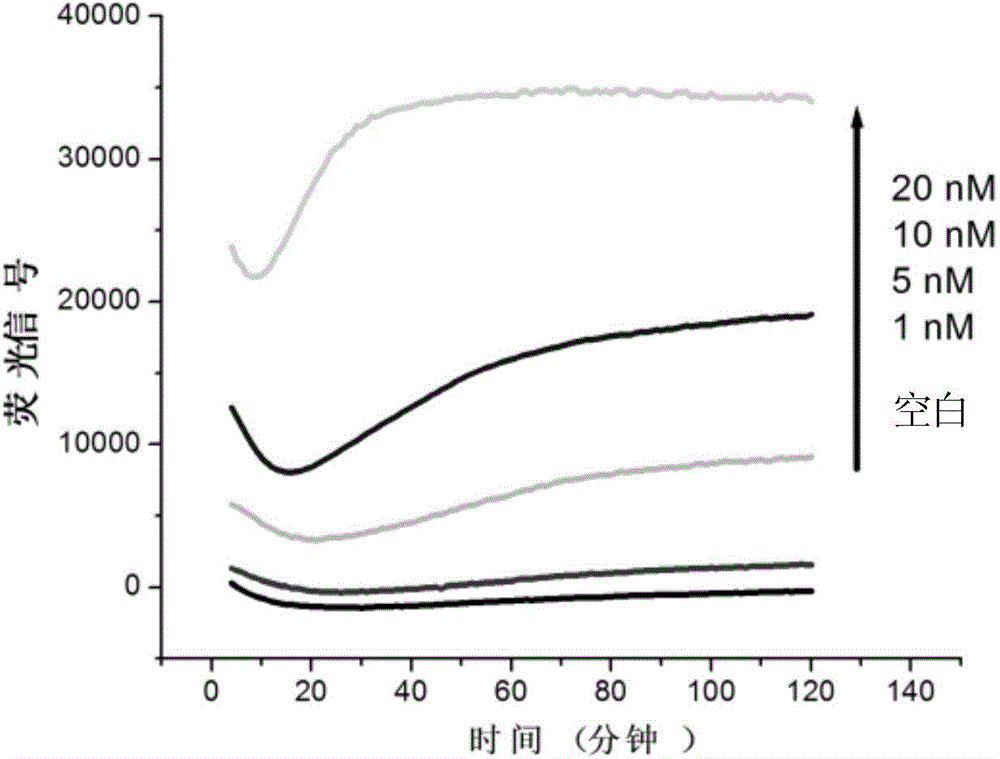
[0019] For the DNA sequence to be tested TAAATGATAGTAGAGTTATGCTAA (provided by Sangon Bioengineering (Shanghai) Co., Ltd.), design a 14-base-length RNA complementary to its base, the base sequence is AUAACUCUACUAUC, and the 3'-end is marked with a black hole quencher group (BHQ1), 5'-terminal labeling 6-carboxyfluorescein (FAM) to obtain a double-labeled fluorescence-activated probe (provided by Integrated DNA Technologies (USA)), due to the close distance, the fluorescence signal of FAM is blocked by BHQ1 quenched. Add double-labeled fluorescent-activated probes, RNase H, and DNA standard samples to be tested in 1×RNase H buffer medium, mix well, and react at 37°C, and control the initial concentration of double-labeled fluorescent-activated probes in the reaction system as 0.8μmol / L, the initial activity of RNase H is 2.5U, and the initial concentration of the DNA standard sample to be tested in the reaction system is controlled to be 0, 1, 5, 10, 20nmol / L, and the reaction ...
Embodiment 2
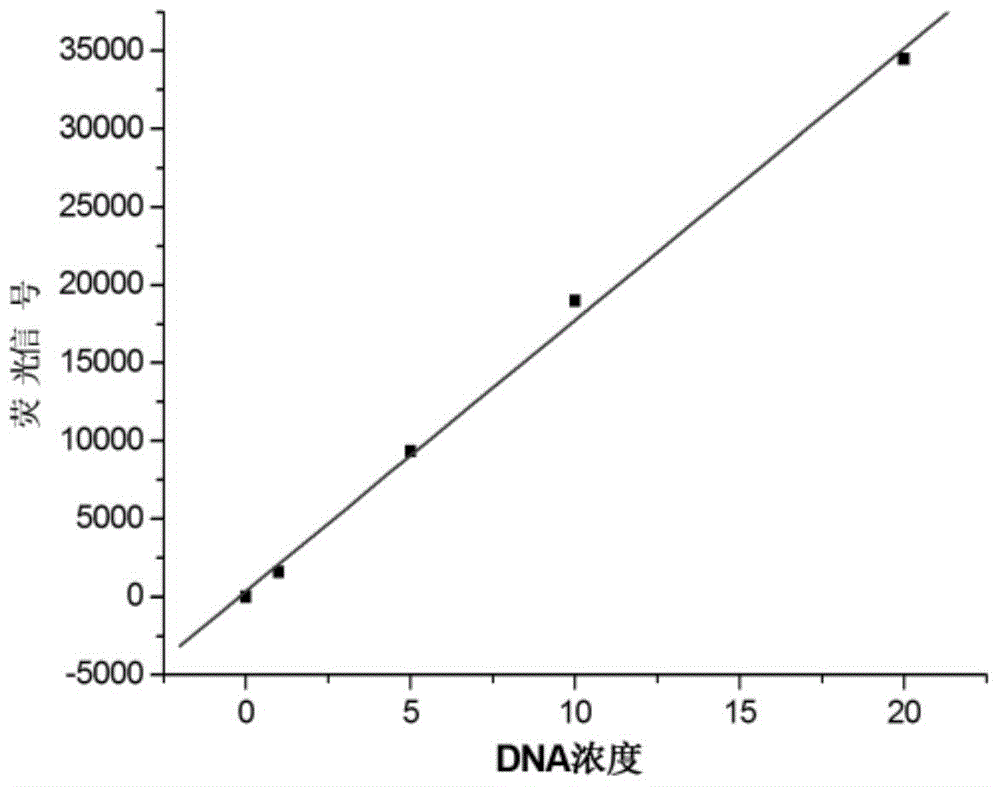
[0024] For the DNA sequence to be tested TAAATGATAGTAGAGTTATGCTAA (provided by Sangon Bioengineering (Shanghai) Co., Ltd.), design a DNA with a length of 14 bases complementary to its base, and wherein 3 consecutive bases are RNA bases. The base sequence is ATAAC wxya ACTATC, the 3'-end was labeled with BHQ1, and the 5'-end was labeled with FAM to obtain a double-labeled fluorescence-activated probe (provided by Integrated DNA Technologies (USA)). Due to the close distance, the fluorescence signal of FAM was quenched by BHQ1. According to the method of embodiment 1, first detect that fluorescence intensity changes with DNA standard sample concentration (C DNA ) changes in the fluorescence spectrum (see Figure 4 ), and draw a linear curve of the fluorescence intensity changing with the concentration of the DNA standard sample to be tested after 120 minutes of reaction (see Figure 5 ),Depend on Figure 4 It can be seen that as the concentration of the DNA standard sample t...
Embodiment 3
[0028] In addition to the direct detection of DNA molecules in biological samples, the double-labeled fluorescence-activated probe of the present invention can also be used for the detection of DNA products in nucleic acid amplification systems. The following uses the analysis of RCA products as an example. The principle is as follows: Image 6 As shown, the specific implementation steps are as follows:
[0029] 1. For a mutated DNA sequence TTTGTGCCTGTCCTGGGAGAGACTGGCGCACAGAGGAAGAGAATCT (provided by Integrated DNA Technologies (USA)) for exon 8 of human p53 gene, design a padlock probe GTCTCTCCCAGGACAGGCTTTTGATCACAGTTTACGGTTTAGCATAACTCTACTATCATTTACTTTACGATTTCGGCTCTTCCTCTGTGCGCCA (provided by DNAAnoTech from Integrated DNATech) Add padlock probes, ligase, and DNA standard samples to be tested in the ligase buffer medium, mix well, and react at 70°C, control the initial concentration of padlock probes in the reaction system to 20nmol / L, and the initial activity of ligase 1.0U, an...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com