RNA induced silencing complex mediated shearing site and uses thereof
A site and DNA molecule technology, applied in the field of cut sites, can solve problems such as difficult to predict the real high-level structure of viral RNA, application barriers, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
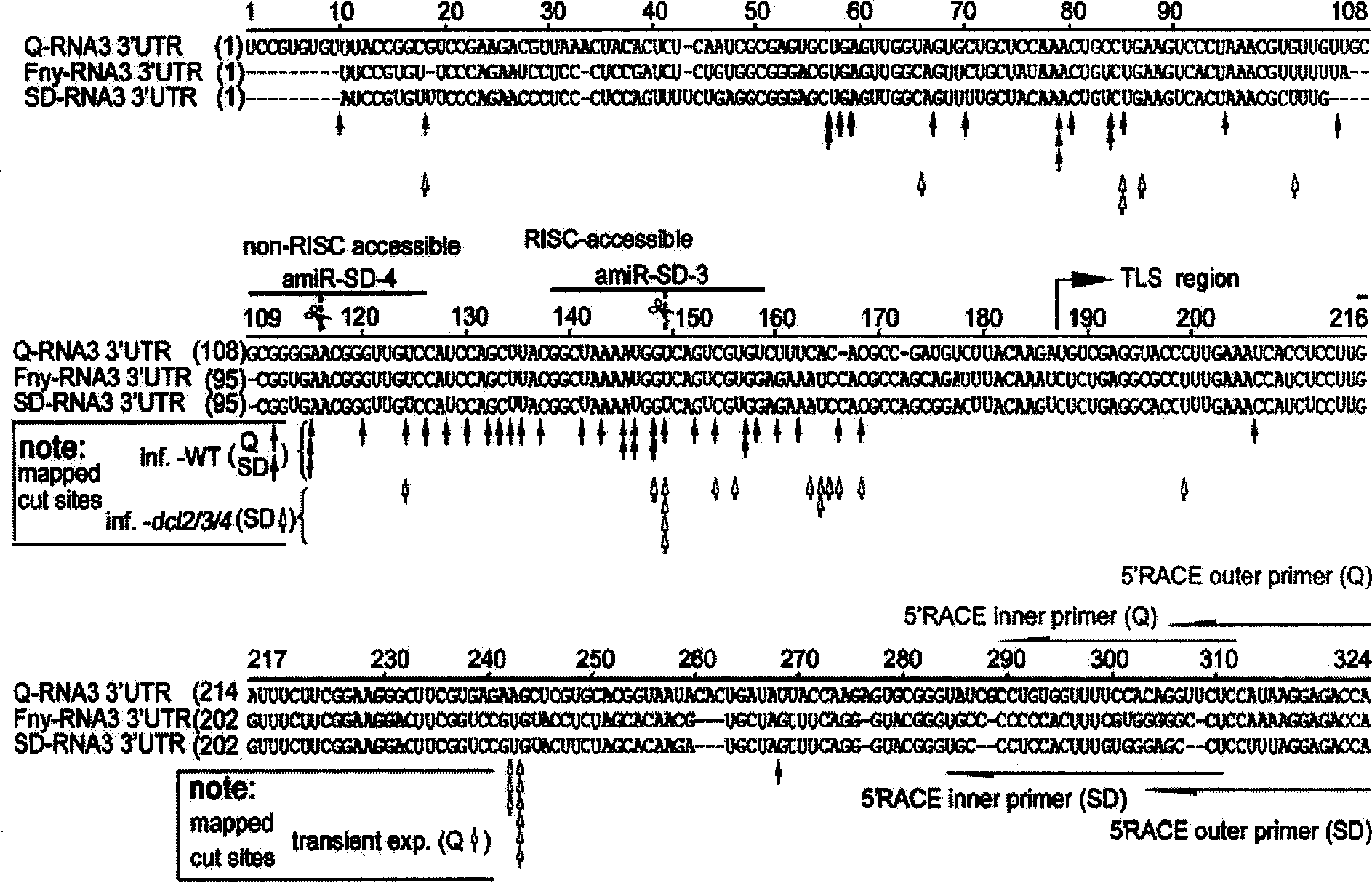
[0032] Embodiment 1, the design of the amiRNA targeting the 2051 / 2052 site in the 3'UTR region of the cucumber mosaic virus genome
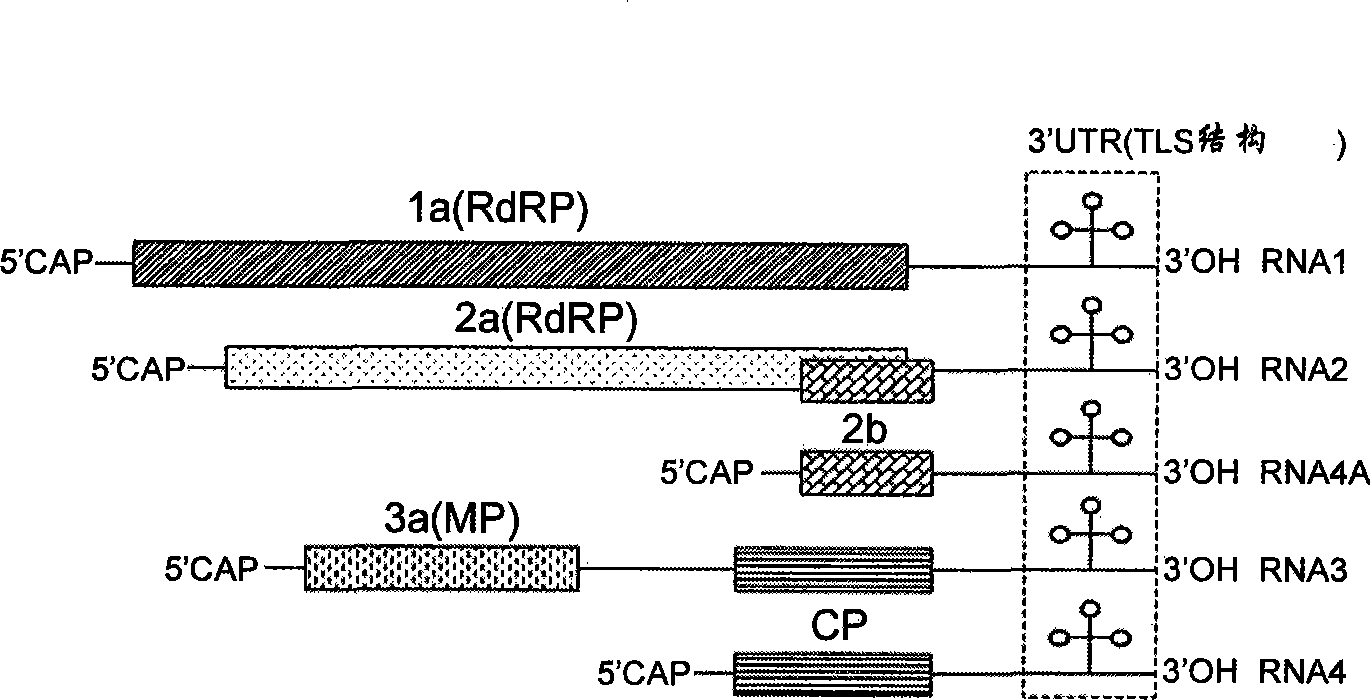
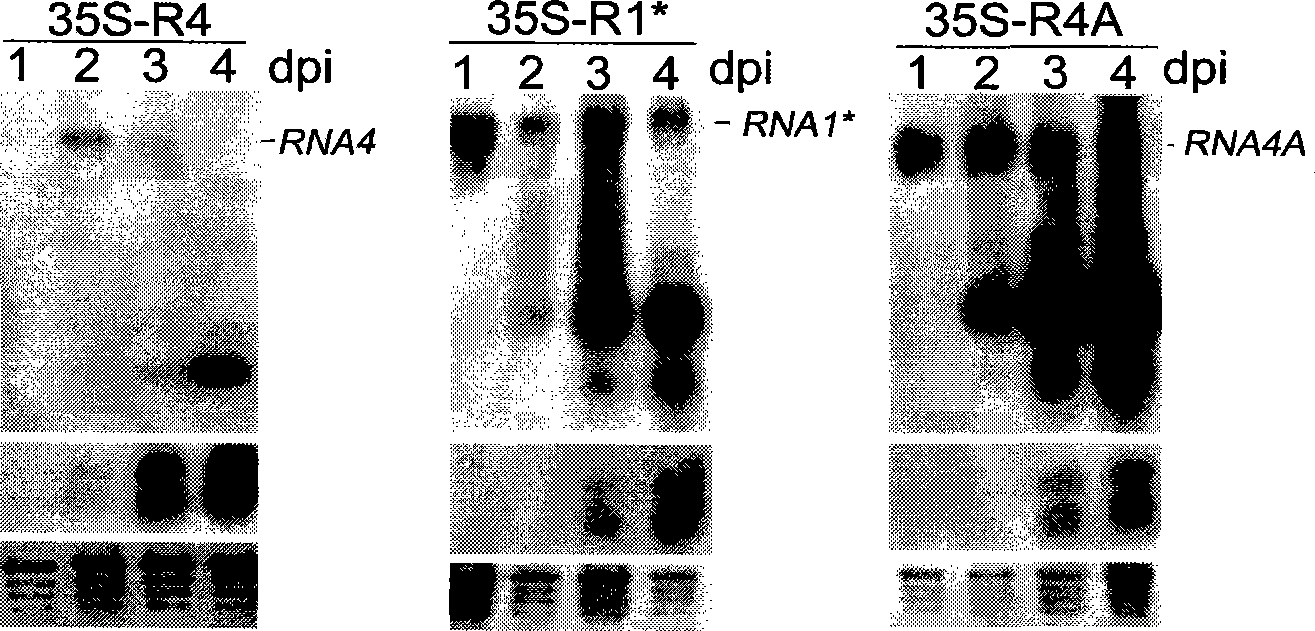
[0033] 1) Cucumber mosaic virus (CMV) 3′UTR can be recognized by plant endogenous RNA silencing pathway
[0034] In order to study whether the 3′UTR region can be recognized and targeted for degradation by the plant RNA silencing system during CMV infection, in view of the fact that the CMV genome is too long, in order to better maintain the natural folding structure of the 3′UTR region, a Q-CMV Expression vectors 35S-R4A and 35S-R4 for complete subgenomic RNA4A and RNA4. The construction method of expression vector 35S-R4A and 35S-R4 is as follows: pQCD2 (Ding, S.W., J.P.Rathjen, W.X.Li, R.Swanson, H.Healy, and R.H.Symons.1995.Efficient infection from cDNA clones of cucumber mosaic cucumovirus RNAs in a new plasma vector.J.Gen.Virol.76(Pt2):459-464) (Institute of Microbiology, Chinese Academy of Sciences) as a template with QR4-5(5'-TCTAGAAGCGT...
Embodiment 2
[0048] Embodiment 2, cultivating transgenic plants resistant to cucumber mosaic virus with the amiRNA of embodiment 1
[0049] One, the amiRNA of embodiment 1 cultivates the transgenic plant resistant to cucumber mosaic virus
[0050] a) Construction of amiRNA precursor with miR159a precursor as the backbone
[0051] The Arabidopsis miR159a precursor 266bp fragment was amplified by RT-PCR using the Arabidopsis genomic DNA as a template. In addition to the complete pre-miR159a (184bp), the 5′ end of the fragment also contained 82bp genome sequence. The sequences of primers miR159-For and miR159-Rev used in RT-PCR are as follows:
[0052] miR159-For: 5′-TCTAGACCACAGTTTGCTTATGTCGGATCC-3′ and miR159-Rev: 5′-ATG TAGAGCTCCCTTCAATCC -3'.
[0053] The italic part at the 5' end of the miR159-For primer is the introduced XbaI site, and the underlined part of the miR159-Rev primer is 18 nucleotides in the 21 nucleotide sequence of the mature miR159.
[0054] The fragment obtained a...
Embodiment 3
[0110] Specificity analysis of the amiRNA of embodiment 3, embodiment 1 in transgenic plants
[0111] In order to exclude the non-specific silencing of plant endogenous genes by the stable expression of amiRNA transgenes, that is, the so-called "off-target" phenomenon, the sequences of all amiRNAs were entered into the NCBI BLAST website for comparison and analysis. It was found that in the 21nt sequences of amiRNAs, there was no Any amiRNA can pair with the continuous complementarity of Arabidopsis endogenous transcripts beyond 15nt. In order to further exclude the possibility of amiRNA targeting plant endogenous genes, at the same time input the amiRNA sequence into "Plant miRNA Target Finder"
[0112] ( http: / / bioinfo3.noble.org / miRNA / miRU.htm ) "TIGR Arabi dopsis GeneIndex 1" and "TIGR Tobacco Gene Index 1" databases for potential target gene searches. The results showed that only one endogenous transcript - AT3G29250 - could form less than 3 incomplete pairs with amiR-...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com