Device and method for genealogy reestablishing on molecular level
A technology of molecular level and genealogy, which is applied in special data processing applications, instruments, electrical digital data processing, etc., can solve the problem that the genealogy at the plant molecular level cannot be reconstructed, and achieve the effect of fast genealogy reconstruction and simple operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
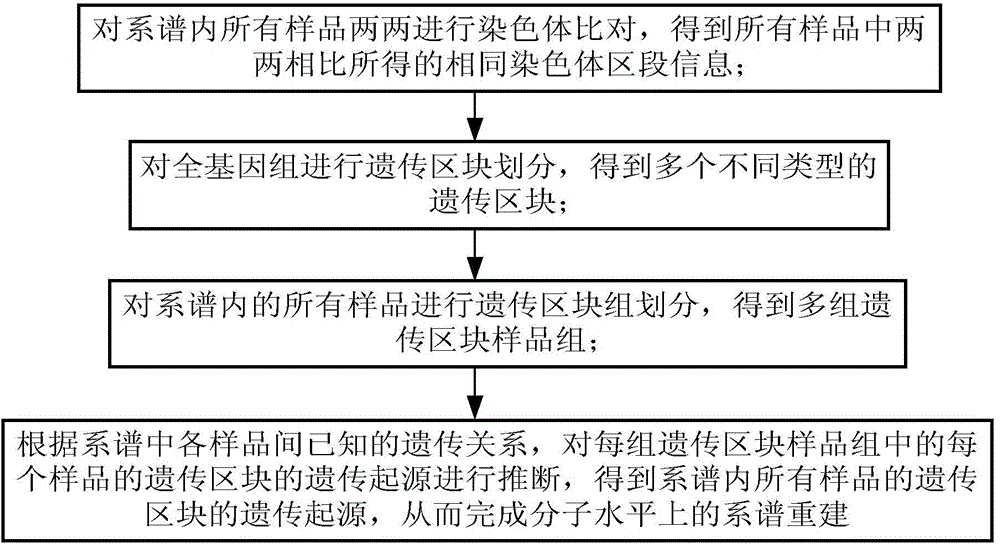
[0066] The object of embodiment 1 is an orthologous pedigree composed of 7 samples. It is known that sample A3 is the descendant generation of samples A1 and A2, sample A3 and sample A4 are the parent generation of A5, and the descendant generation samples of samples A5 and A6 For A7, the high-throughput sequencing data of 7 samples in the pedigree all exist, as follows figure 1 The shown pipeline reconstructs this family tree at the molecular level:
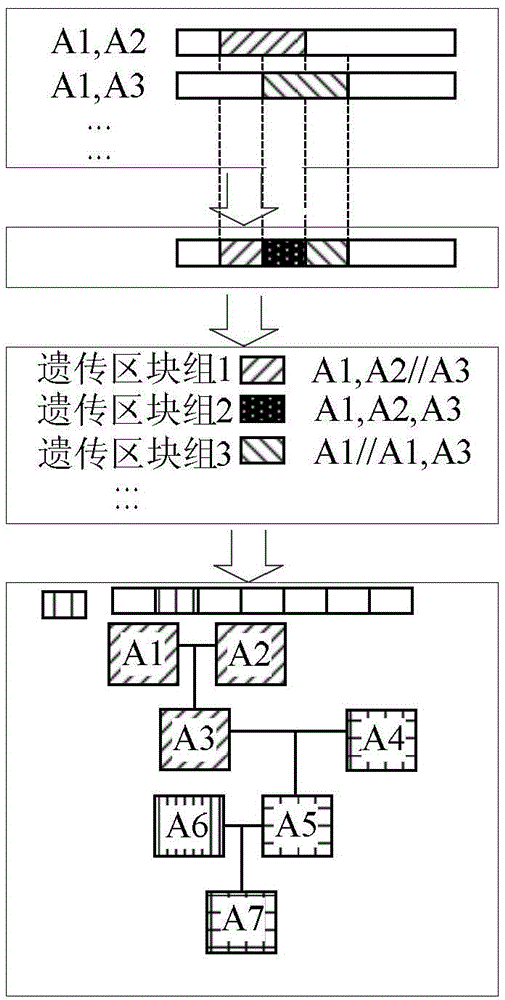
[0067] The IBDseq algorithm is used to compare the IBD block information of the 7 samples in the pedigree, such as figure 2 As shown, sample A1 and sample A2 are subjected to IBD block detection to obtain the chromosome information shared between sample A1 and sample A2; similarly, A1 and sample A3 are then subjected to IBD block detection to obtain sample A1 and sample A3 Chromosome information shared between, as mentioned above, A1 and A4, A1 and A5, A1 and A6, A1 and A7, A2 and A3, A2 and A4, ... and A6 and A7, that is, all...
Embodiment 2
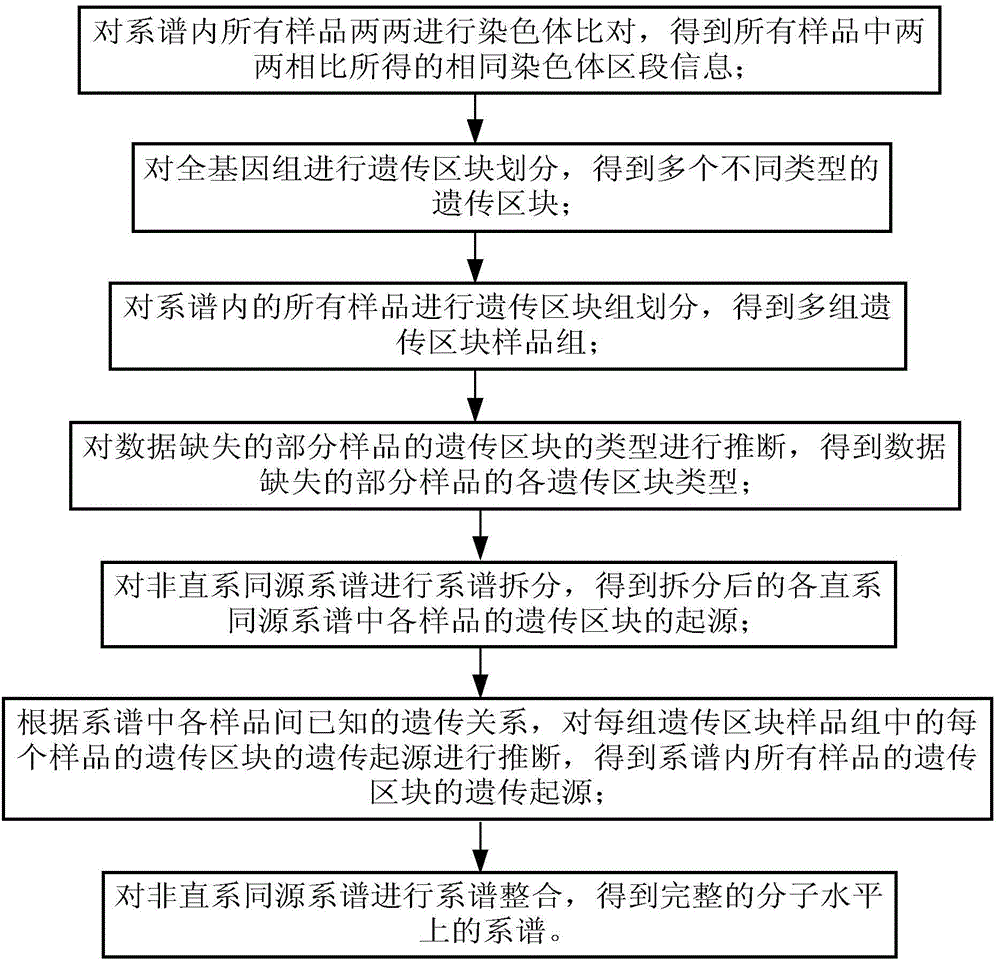
[0073] The object of embodiment 2 is a pedigree composed of 9 samples belonging to different orthologous pedigrees. It is known that sample A3 is the offspring generation of samples A1 and A2, sample A3 and sample B3 are the parent generation of A5, and sample A5 The offspring sample of A6 and A6 is A7, and the parents of sample B3 are B1 and B2; the high-throughput sequencing data of sample A6 is missing. follow below image 3 The shown pipeline reconstructs this family tree at the molecular level:
[0074] The steps of carrying out IBD block detection, genetic block division and grouping of 9 samples according to genetic block types are the same as in Example 1 for the 9 samples in the pedigree;
[0075] In order to ensure the continuity of the pedigree and the integrity of the data, it is necessary to infer the most likely genetic block type of sample A6 with missing data according to the genetic block type of close relatives:
[0076] When the genetic block types of the ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com