Method for preparing exogenous circular RNA and method for quantitatively determining endogenous circular RNA
An exogenous circular and exogenous technology, applied in the field of molecular biology, can solve problems such as lack of circular RNA reference genes
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
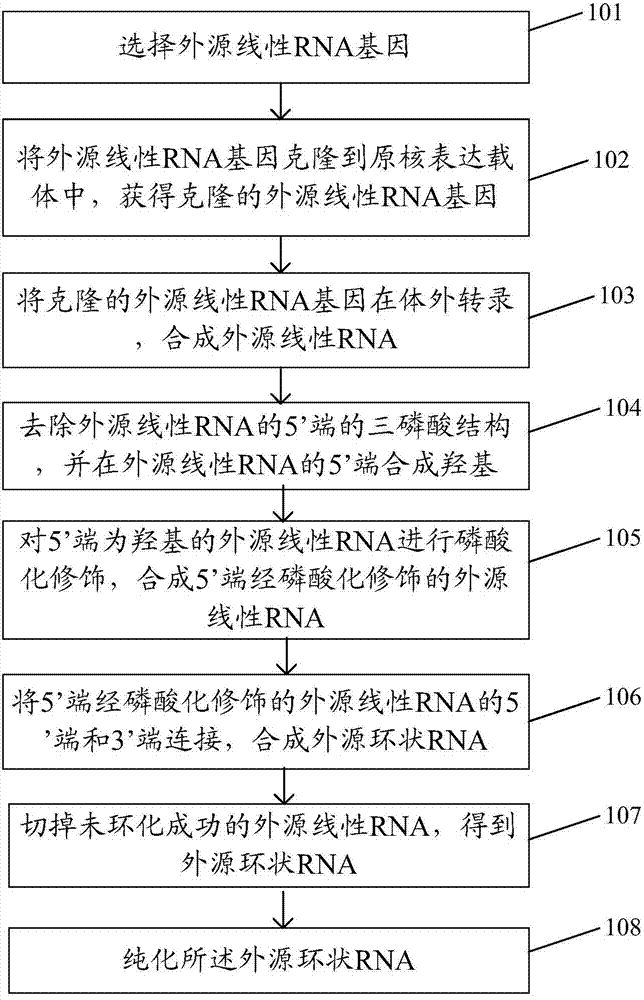
[0044] The embodiment of the present invention provides a method for preparing exogenous circular RNA, such as figure 1 As shown, the method includes:
[0045] Step 101: selecting an exogenous linear RNA gene;
[0046] Step 102: Cloning the exogenous linear RNA gene into a prokaryotic expression vector to obtain the cloned exogenous linear RNA gene;
[0047] Step 103: Reverse transcribing the cloned exogenous linear RNA gene in vitro to synthesize exogenous linear RNA;
[0048] Step 104: removing the triphosphate structure at the 5' end of the exogenous linear RNA, and synthesizing a hydroxyl group at the 5' end of the exogenous linear RNA;
[0049] Step 105: performing phosphorylation modification on the exogenous linear RNA whose 5' end is hydroxyl, and synthesizing the exogenous linear RNA with phosphorylation modification at the 5' end;
[0050] Step 106: linking the 5' and 3' ends of the phosphorylated exogenous linear RNA at the 5' end to synthesize exogenous circular...
Embodiment 2
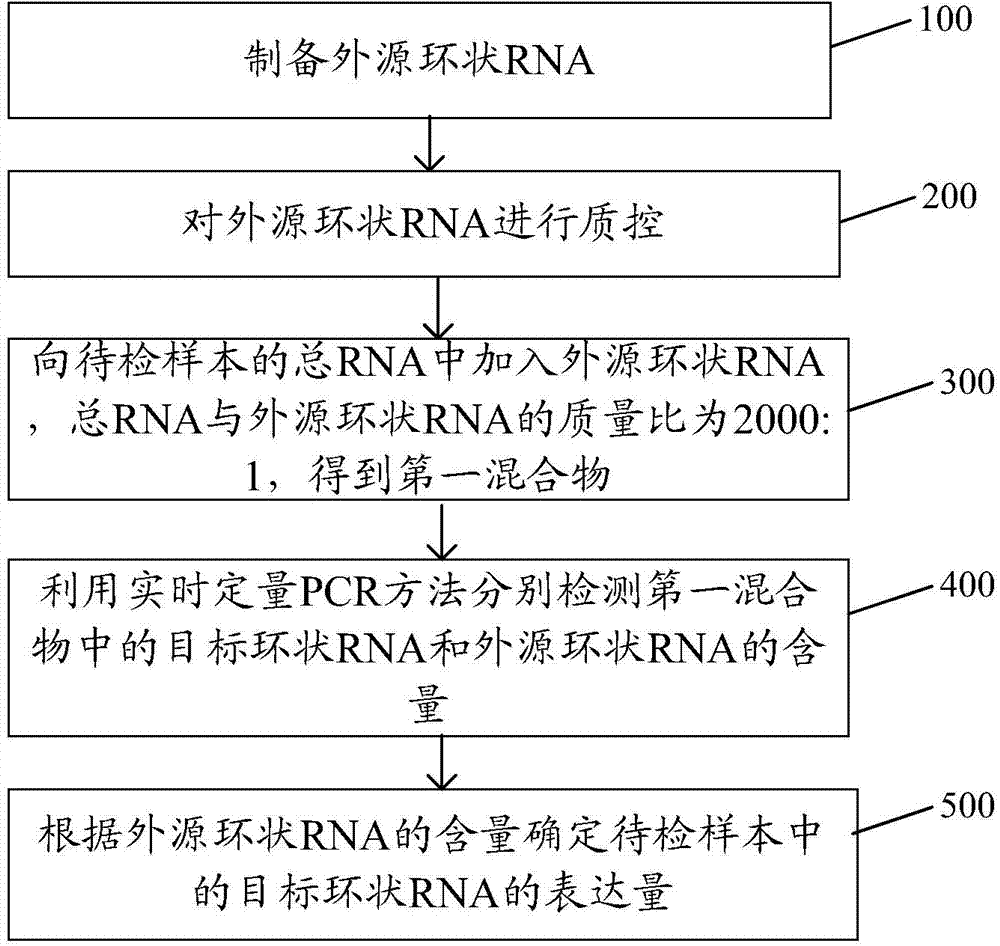
[0077] The embodiment of the present invention provides a method for quantitative detection of circular RNA, such as figure 2 As shown, the method includes:
[0078] Step 100: Using the method provided in Example 1 of the present invention to prepare exogenous circular RNA.
[0079] Step 200: performing quality control on the exogenous circular RNA.
[0080] Step 300: adding exogenous circular RNA to the total RNA of the sample to be tested and the control sample, the mass ratio of total RNA to exogenous circular RNA is 2000:1, to obtain a mixture of exogenous circular RNA and total RNA.
[0081] Step 400: Using real-time quantitative PCR to detect the contents of the target circular RNA and the exogenous circular RNA in the mixture of the exogenous circular RNA and the total RNA respectively.
[0082] Step 500: Using the exogenous circular RNA as a reference gene, determine the relative expression level of the target circular RNA in the sample to be tested.
[0083] Where...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com