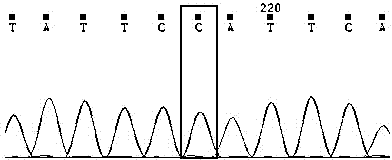
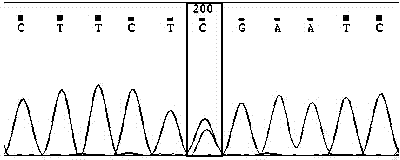
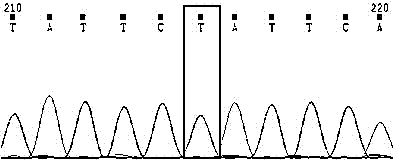
Method for verifying anti-chalk disease traits of swarm by SNP marker
A chalk disease and bee colony technology, applied in the field of using SNP markers to identify bee colony resistance to chalk disease, can solve the problems of less research and utilization, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0035] A method utilizing SNP markers to identify anti-chalk disease traits of Apis mellifera colony, comprising the following steps:
[0036] (1) Sampling of bee larvae: 3-day-old honeybee larvae were randomly collected from the Italian bee colonies without other obvious diseases to be investigated, 20 bee larvae per group, each individual was placed in a centrifuge tube, and then Freeze at -20°C for later use;
[0037] (2) Extraction of DNA from larval samples (using Sangon’s Animal Genome Rapid Extraction Kit, SK8222)
[0038] Place the larvae from step (1) in a centrifuge tube for tissue disruption, add 350 ul Buffer Digestion, shake evenly, and place in a water bath at 65 °C for 1 h until the cells are completely lysed.
[0039] Add 150 ul Buffer PA, mix thoroughly by inverting, and place in a -20°C refrigerator for 5 min.
[0040] Centrifuge at 10,000 rpm at room temperature for 5 min, transfer 400 ul of the supernatant to a new 1.5 ml centrifuge tube, if the supe...
Embodiment 2
[0076] The specific data are as follows:
[0077] Table 1A The effect of different genotypes on the disease resistance of bee colonies and the distribution of gene frequency
[0078]
[0079] Note: Chi-square test of the effect of different genotypes on the disease resistance of bee colonies P < 0.05.
[0080] Table 1B The effect of different genotypes on the disease resistance of bee colonies and the distribution of gene frequency
[0081]
[0082] Note: Chi-square test of the effect of different genotypes on the disease resistance of bee colonies P < 0.05.
[0083] Table 1C The effect of different genotypes on the disease resistance of bee colonies and the distribution of gene frequency
[0084]
[0085] Note: Chi-square test of the effect of different genotypes on the disease resistance of bee colonies P < 0.05.
[0086] (1) The statistical results of Table 1A, Table 1B, and Table 1C through Fisher's Exact Test (Fisher's Exact Test) show that there are s...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com