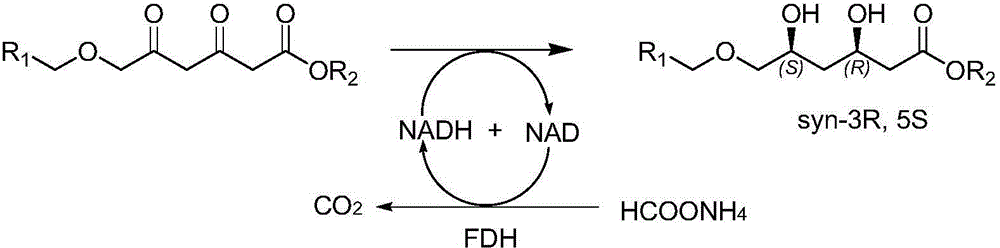
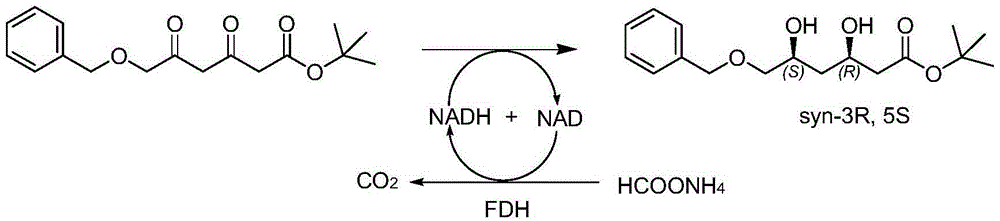
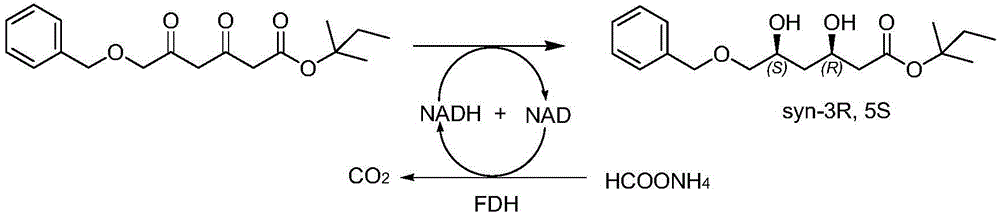
Diketoreductase mutant and application thereof
A technology of reductase and dicarbonyl, which is applied in the field of mutants of dicarbonyl reductase, can solve the problems of large amount of enzyme solution, increased total volume of reaction system, low enzyme catalytic activity, etc., and achieves the improvement of enzyme activity and enzyme specificity. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0056] Site-directed saturation mutation was performed on a double carbonyl reductase (DKR) mutant derived from Rhodococcus erythropolis SK121 strain (the amino acid sequence of which is shown in SEQ ID NO: 6).
[0057] The amino acid sequence of double carbonyl reductase (DKR) was simulated on the Swiss-model website to simulate the three-dimensional structure of the protein, and then the binding simulation between the substrate and the protein was carried out through Docking. Finally, through Pymol analysis, the selection may be related to the binding of the substrate and NAD. Amino acids related to NAD proton transport were used as mutant amino acids ( Figure 4 ).
[0058] According to the mutated amino acid and the base sequence on both sides (see the mutation site in Table 1 for the mutated amino acid), use Primmer 5.0 to design the corresponding mutant primers (Table 1). Using the pET22b(+) expression vector containing the double carbonyl reductase gene (purchased from...
Embodiment 2
[0062] Example 2: Preliminary screening of double carbonyl reductase mutants
[0063] According to the content described in Example 1, pick a single colony on the above-mentioned solid medium and inoculate it in a 96-deep well plate, add 1 ml of LB liquid medium containing 50 μg / ml ampicillin in advance to each well, and culture at 37°C with shaking at 220 rpm After 3 hours, add IPTG at a final concentration of 0.1 mM, induce culture at 18°C, 220 rpm for 16 hours, and centrifuge at 4000 g for 15 minutes to collect the cells, and the cells are broken by an ultrasonic breaker (JY92-2D, Ningbo Xinzhi Biotechnology Co., Ltd.), 4 Centrifuge at 12000rpm for 5min to obtain the supernatant, that is, the crude enzyme solution of the mutant, which is used for primary activity screening with a microplate reader. Add 30 μL DMSO, 1.5 μL main material tert-butyl 6-benzyloxy-3,5-dioxo-hexanoate (30 mg / mL dissolved in DMSO), 2.5 μL NADH (20 mg / mL) to the 96-well plate, 216 μL of phosphate bu...
Embodiment 3
[0070] Example 3: Re-screening of double carbonyl reductase mutants
[0071] Inoculate the mutant whose enzyme activity is higher than that of the parent in Example 2 into 500 ml of LB liquid medium containing 50 μg / ml ampicillin, culture with shaking at 37° C. until OD600=0.6, add IPTG to a final concentration of 0.1 mM, and Expression was induced at 18°C. After 16 hours of induction, the cells were collected by centrifugation at 6000g for 10 minutes. The cells were disrupted with an ultrasonic disruptor (JY92-2D, Ningbo Xinzhi Biotechnology Co., Ltd.), and centrifuged at 10,000 g for 20 min at 4°C to obtain the supernatant, which was used for activity detection. Add 0.05g of the main material (tert-butyl 6-benzyloxy-3,5-dioxo-hexanoate or neopentyl 6-benzyloxy-3,5-dioxo-hexanoate to a 10ml reaction flask ), 0.5ml polyethylene glycol PEG-400, after the raw materials are dissolved, add 4.0ml phosphate buffer (100mM, pH=6.0), the main raw material is evenly dispersed in the b...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com