Polymorphic primers for Bactrian camels and their screening methods and methods for identifying parent-child relationship
A polymorphic primer, parent-child relationship technology, applied in biochemical equipment and methods, microbial determination/inspection, DNA/RNA fragments, etc., can solve problems such as heavy workload and easy loss, and achieve high polymorphism, The effect of broad application prospects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0040] This embodiment provides a polymorphic primer for microsatellite molecular markers in the Bactrian camel transcriptome and a deletion method thereof.
[0041] 1. Polymorphic primer screening of Bactrian camel transcriptome microsatellite molecular markers
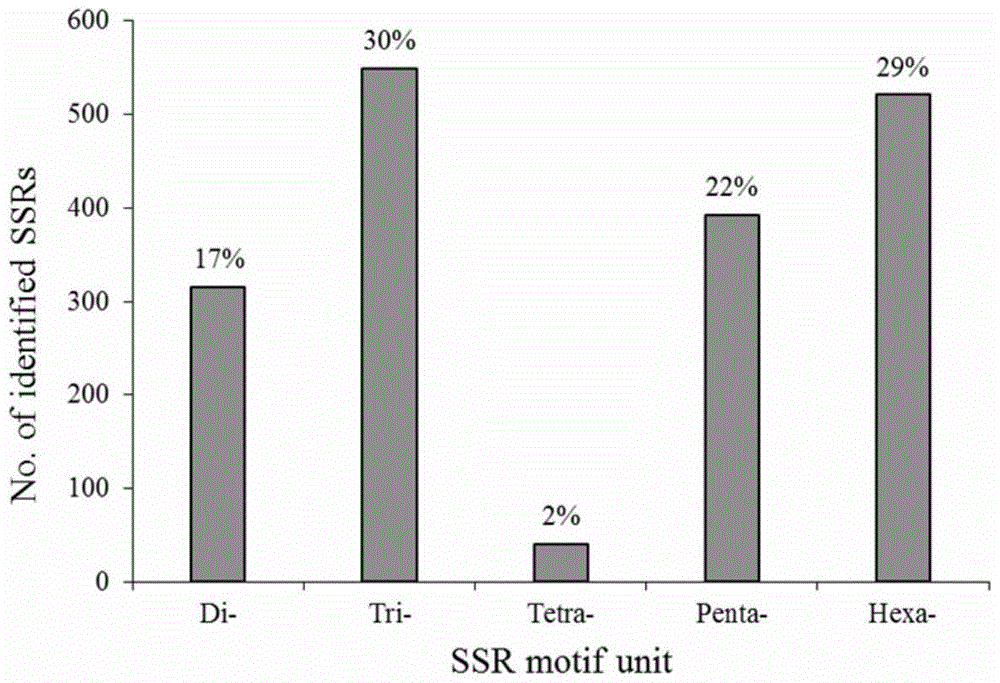
[0042] The transcriptome data of wild Bactrian camels were downloaded from the NCBI public database, and a total of 23,624 transcriptome sequences were obtained, which were independently stored in the FASTA format. Use MIcroSAtellite (MISA http: / / pgrc.ipk-gatersleben.de / misa / misa.html) to analyze the Bactrian camel transcriptome sequence, and screen the Bactrian camel microsatellite molecular sequence. The screening criteria are: repeats of two bases, three bases, four bases, five bases and six bases are respectively 7, 6, 5, 4, 3 and 2 (single nucleotide repeats are not selected because It is generally considered that it is not a useful polymorphic marker); a total of 1820 SSR sites were obtained, and the ratio of ...
Embodiment 2
[0063] The present embodiment provides a method for identifying the parent-child relationship of Bactrian camels using the polymorphic primers of the Bactrian camel transcriptome microsatellite molecular markers provided in Example 1, specifically as follows:
[0064] First use cervus 3.0 software to count the PE1 (non-parent exclusion rate: the genotypes of both parents are known), PE2 (non-father exclusion rate: only know the genotype of the father or mother) of the 117 Bactrian camel samples in Example 1 and the total non-parent exclusion probability (PE total ), 117 Bactrian camel samples PE1, PE2 and PE total See Table 5 for details. PE1, PE2, and PE total Calculation reference formula (Jamieson A. & Taylor S.C. Comparisons of three probability formula for parentage testing, Animal Genetics, 1997, 28, 397-400):
[0065] Paternity Exclusion Rate (PE1):
[0066]
[0067] Paternity Exclusion Rate (PE2):
[0068]
[0069] The overall paternal exclusion rate (PE to...
Embodiment 3
[0093] This embodiment provides a polymorphic primer of 14 pairs of Bactrian camel transcriptome microsatellite molecular markers provided in Example 1 to analyze the genetic diversity of Bactrian camels for revealing the genetic diversity of Bactrian camel samples Current situation, based on 14 pairs of polymorphic primers of microsatellite molecular markers in the Bactrian camel transcriptome and the diversity information detected in 117 Bactrian camel samples, the diversity analysis of each population was performed using PopGene1.32 software, in which:
[0094] Calculate the main genetic diversity parameters and observe heterozygosity (observed heterozygosity, Ho);
[0095] Expected heterozygosity (expected heterozygosity, He);
[0096] Number of alleles (number of alleles, N A );
[0097] Effective number of alleles (Ne);
[0098] Polymorphism information content (polymorphism information index, PIC) calculation reference formula (BotsteinD, White RL, Skolnick M, Davis ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com