NTR1 micromolecular antagonist
A technology of small molecules and antagonists, applied in the field of the structure of NTR1 small molecule inhibitors, can solve the problems of dysregulation of intracellular signal transduction pathways, disordered physiological responses, abnormal physiological phenomena, etc., and achieve the effect of good drug application prospects.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
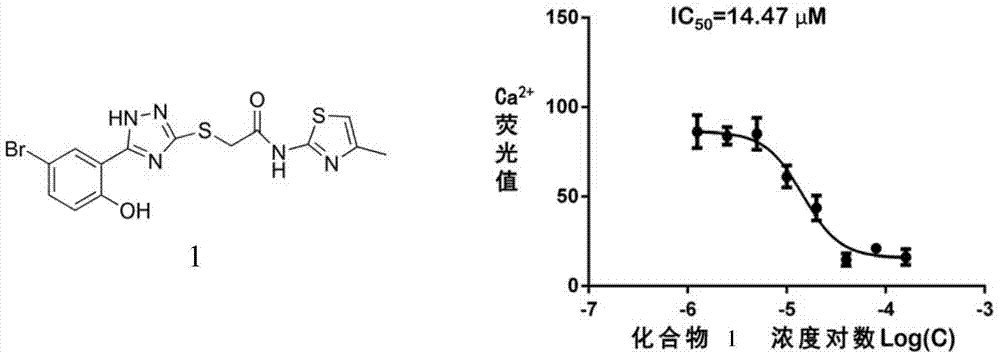
[0020] Example 1, Virtual Screening and Cell Viability Testing of NTR1 Small Molecule Inhibitors
[0021] One, the method for virtual screening of the present invention comprises following 4 steps:
[0022] Step 1: Construction of NTR1 Small Molecule Database
[0023] Integrate multiple commercial small molecule databases (including ZINC, SPECS, J&K Screening Library, InterBioScreening, Timtec, Msdiscove, Iris-biotech, TCM Database, Ambinter Natural Products, AnalytiCon Discovery NP, Princeton NP, Molecular Diversity Preservation International, PUBCHEM), with The Pipeline pilot software removes repetitive molecules and molecules with similar structures, and selects compounds with good absorption and low toxicity according to the calculated ADMET properties of the molecules for Clustering, obtaining a database of 1 million small molecules. The Glide module in Schrodinger was used to dock these small molecules with NTR1 with SP precision, and the top 10,000 small molecules scor...
Embodiment 2
[0039] Example 2. Structural modification based on the mode of action between the small molecule and the receptor NTR1.
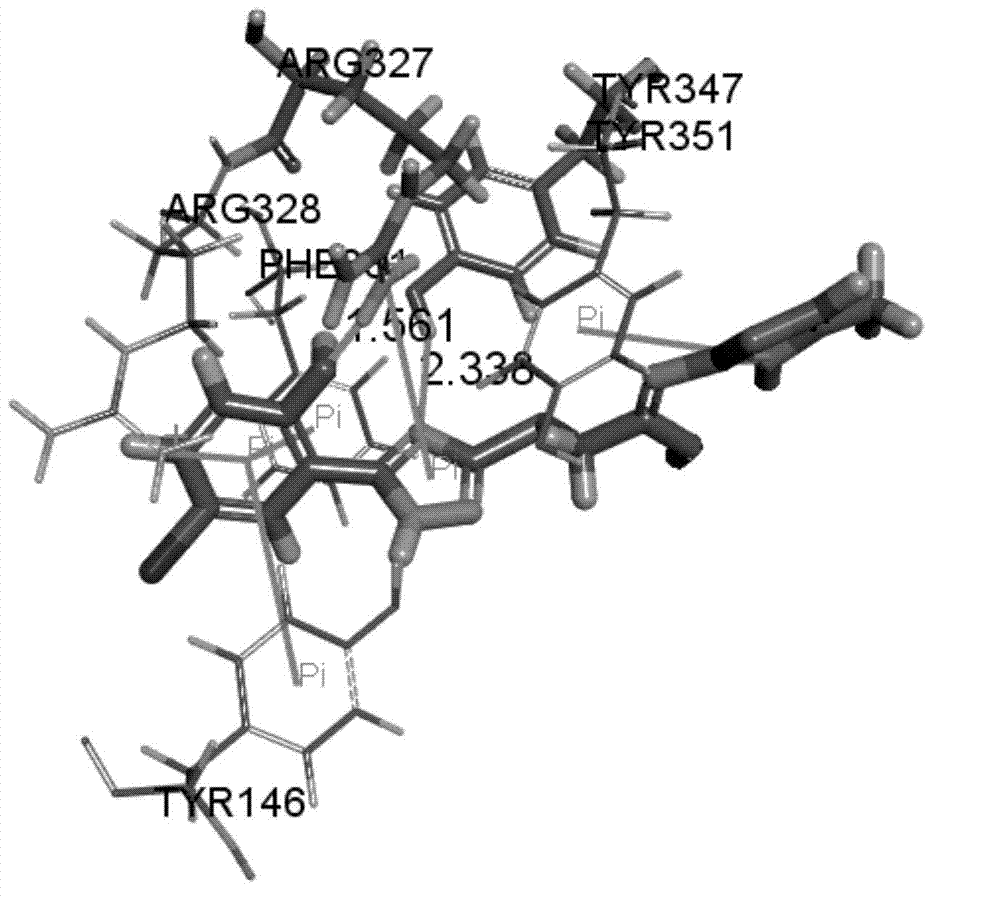
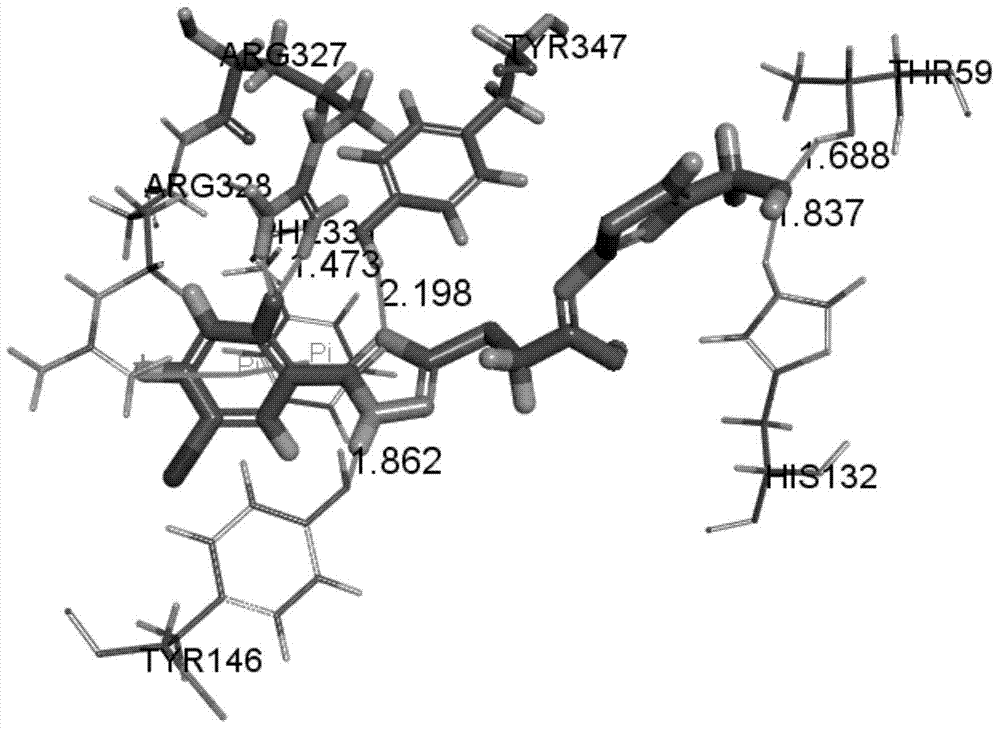
[0040] Firstly, open the three-dimensional crystal structure of NTR1 protein in Discovery Studio3.5, the structure comes from PDB (Protein Data Bank), and the PDB number is 4GRV. Hydrogenate it, remove water, delete the inserted T4 lysozyme structure, use the Prepare Protein module to prepare the protein, and use Define Site to find the active site according to the NT in the ligand. Small molecules were prepared using the Prepare Ligands module. After the preparation is complete, use the Libdock method of the Dock Ligands module for docking, and the docking results are as follows figure 1 shown. According to the docking results, compound 1 forms hydrogen bonds with the key amino acids Arg327 and Tyr347 of the receptor reported in the literature, and the bond lengths of the hydrogen bonds are ( figure 1 ), the hydrogen bonding is very important for the ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com