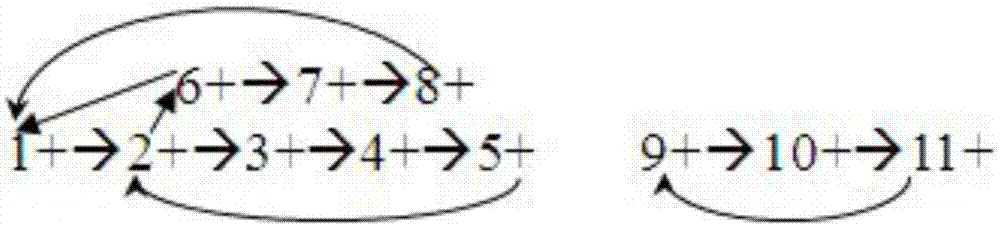Genome sequencing data sequence assembling method
A genome sequencing and reference genome technology, applied in the direction of electrical digital data processing, special data processing applications, instruments, etc., can solve the problem of not being able to accurately determine the exact placement of fragments, and achieve accurate and effective results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
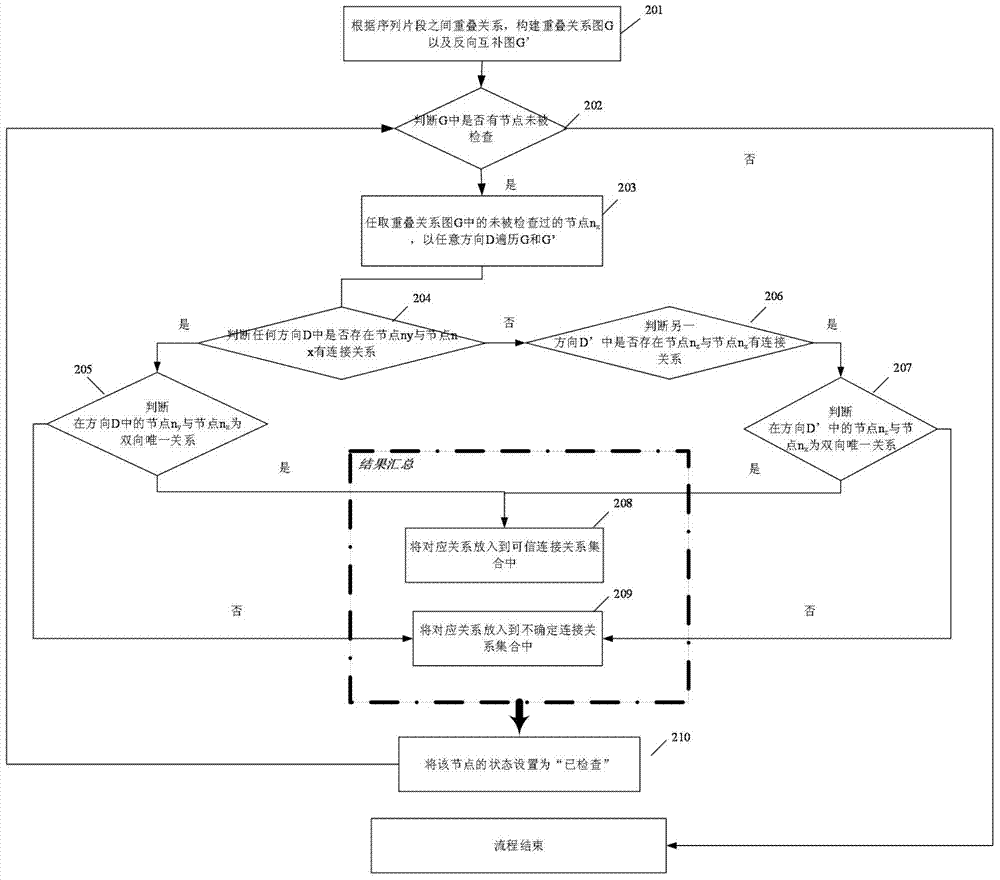
[0034] figure 2 It is a schematic flowchart of the genome sequencing data sequence assembly method in the embodiment of the present invention. like figure 2 As shown, the method includes:
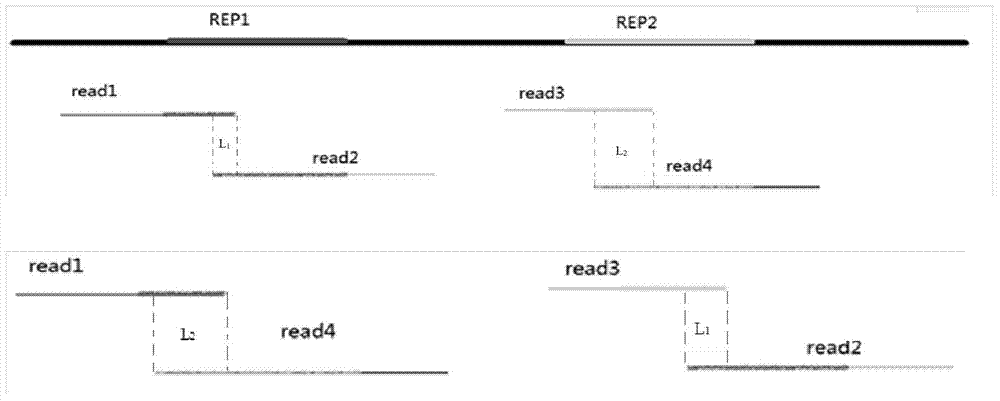
[0035] Step 201: According to the overlap relationship between read sequences obtained by sequencing, construct an overlap relationship graph and a reverse complement graph. All corresponding nodes in the overlapping relationship graph and its reverse complementary graph are in reverse complementary and equivalent relationship with each other. Since we usually only know whether the two sequences have an overlapping relationship, but are not sure about the final arrangement order of the sequence groups in the assembly result, we need to construct two graphs at the same time, the overlapping relationship graph G and its anti-complementary sequence graph G'. As long as there is an overlapping relationship between two sequence fragments, it can be marked in the overlapping relationship dia...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com