Method for developing SSR molecular markers on large scale
A molecular marker, large-scale technology, applied in the fields of molecular biology and bioinformatics, can solve the problems of low effectiveness and high repeatability of SSR marker development, and achieve the effect of improving development efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0043] Specific embodiments of the present invention will be described in detail below.
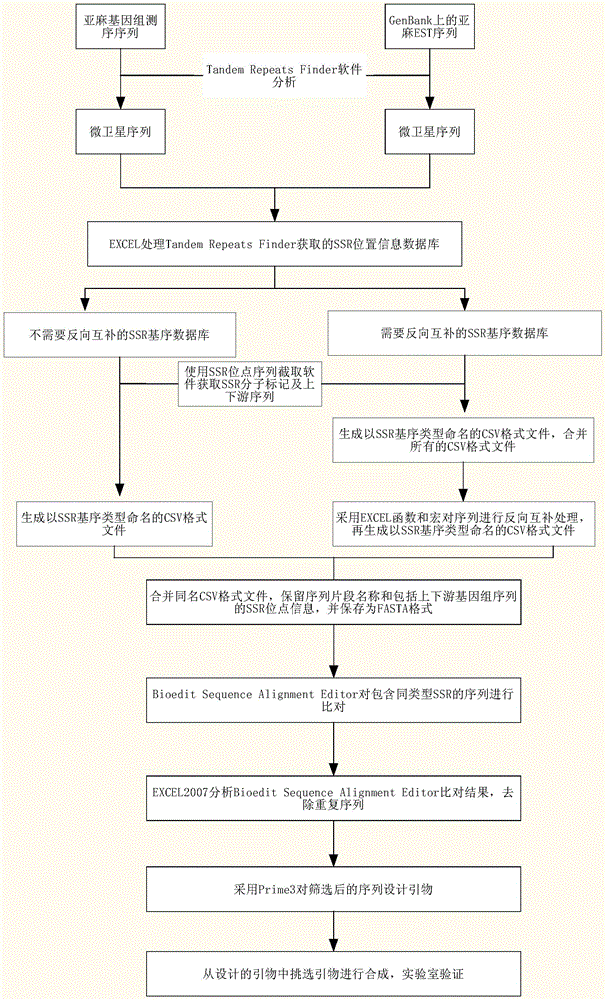
[0044] Such as figure 1 As shown, taking flax as an example, a method for developing SSR molecular markers in large batches includes the following steps:
[0045] (1) Obtain the genome sequencing sequence or the EST sequence on GenBank, and save the genome sequencing sequence or the EST sequence on GenBank in FAS format;
[0046] (2) Use Tandem Repeats Finder to obtain the genome sequencing sequence obtained in step (1) or the SSR site information of the EST sequence on GenBank, and convert the analysis and summary results into a CSV format file, and establish a database 1. CSV, Tandem Repeats The setting parameters of Finder are (20,2000,2,7,7);
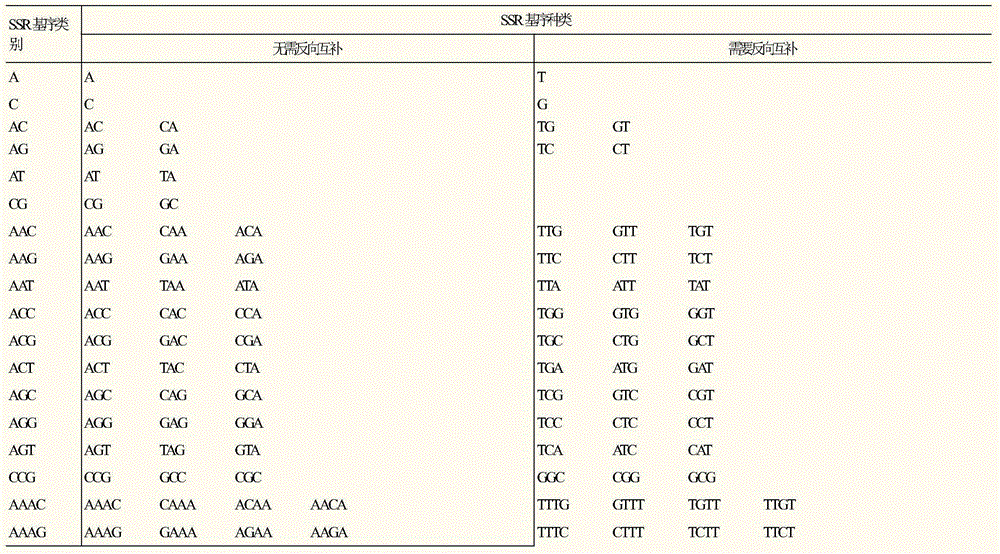
[0047] (3), classify the SSR sites according to the SSR motif type,
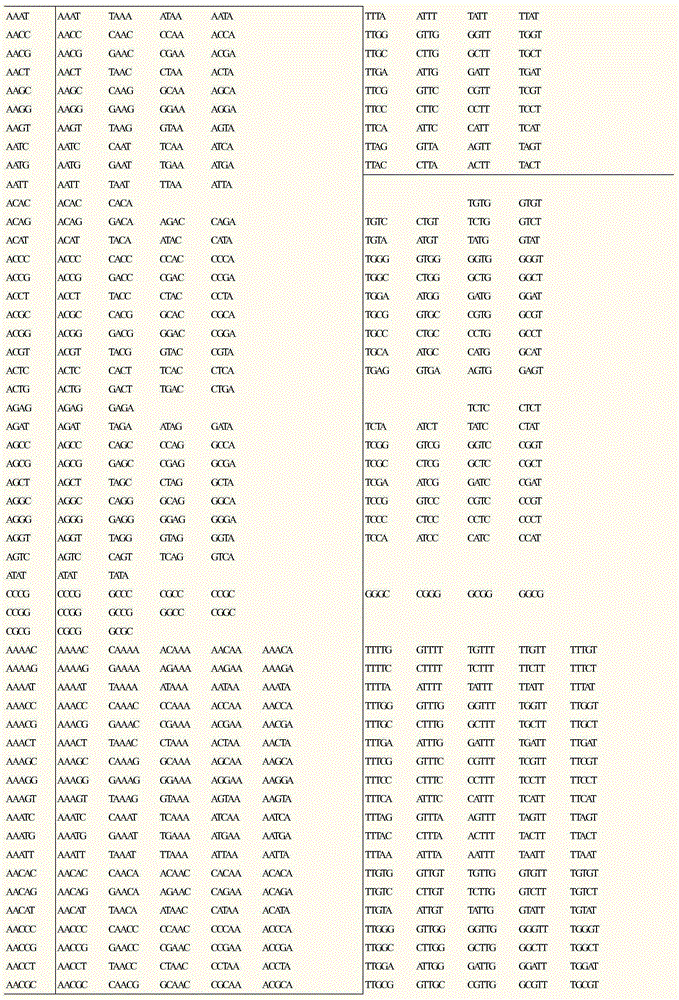
[0048] (31) According to the principle of base complementation and shift combination, all SSR types containing 1 to 6 bases are divided into 508 types, see Table 1 at the ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com