RPA detection-based quarantine solanum weed solanum elaeagnifolium detection method
A silver-haired black nightshade and DNA molecular technology, applied in biochemical equipment and methods, DNA/RNA fragments, recombinant DNA technology, etc., can solve the problems of long time, time-consuming, laborious, and long-term, and achieve high specificity and Stability, simple and fast response, and short response time
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0038] Embodiment 1, the establishment of the preparation of the RPA kit of identification silver-haired nightshade and identification method
[0039] 1. Design of RPA primers for identification of Solanum nigrum
[0040] The present invention is used for the identification kit of the RPA of black nightshade, which contains the primer pair for specific detection of black nightshade, and the recombinase, single-stranded DNA binding enzyme, strand-displacing DNA polymerase, dNTPs, Magnesium acetate, etc. The primer pair design process is roughly as follows:
[0041] According to the nuclear gene Solanumelaeagnifolium voucher Bohs3204UT granule-boundstarch synthase (GBSSI) gene (NCBI: AY996412.1), a quarantine solanum weed, multiple sets of theoretically feasible RPA primers were designed using primer design software, and two sets of primers were selected for specificity experiments Verify that the two sets of primers are:
[0042] Waxy-F1: 5'-AGCTTGTTCTGTCAAGTAAGTTACTAGCTGTAT...
Embodiment 2
[0069] Example 2, Rapid Analysis of RPA
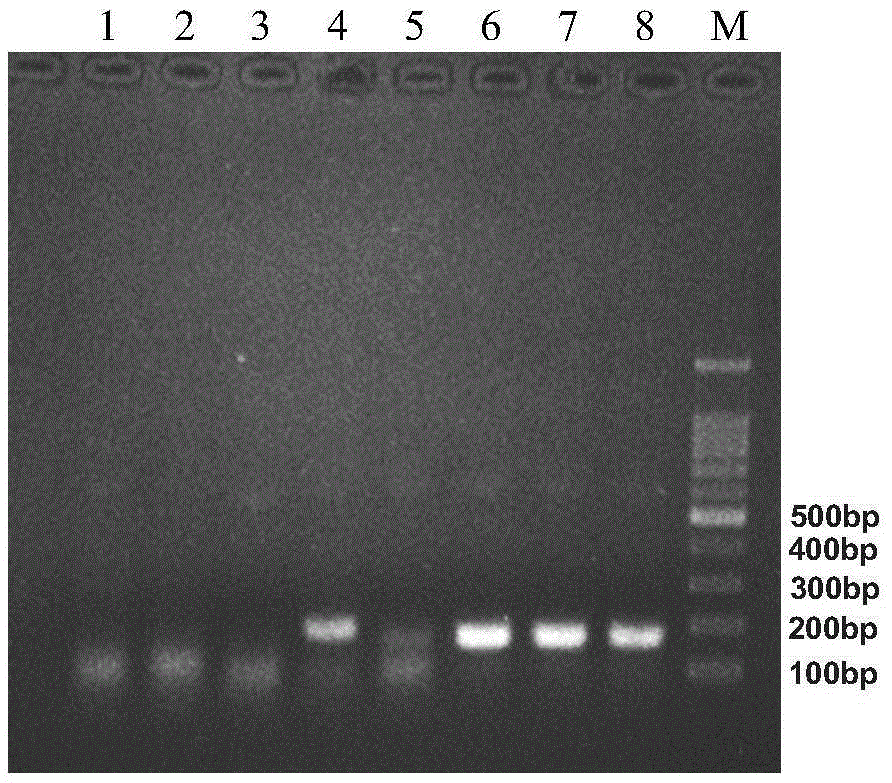
[0070] With the leaves and stems of Solanum nigrum as the test sample, the primer pair (Waxy-F1 / Waxy-R1, sequence 1 and sequence 2) screened and identified in the above-mentioned embodiment 1 is used for RPA detection, and several reaction time gradients are designed, i.e. 10min , 20min, 30min, 40min, 60min, 90min to analyze the influence of RPA reaction time on detection. Refer to Step 2 of Example 1 for the rest of the operations.
[0071] It was found that after 10 minutes of reaction, the target band with a size of 210bp (sequence 3) could be detected. After 20 minutes, the brightness of the amplified fragment did not increase significantly, and after 90 minutes of reaction, the brightness decreased, indicating that the RPA reaction can be performed in a relatively short time. The DNA fragments were amplified within 20 minutes, and this experiment determined that 20 minutes was the optimal time for the RPA reaction. experiment as...
Embodiment 3
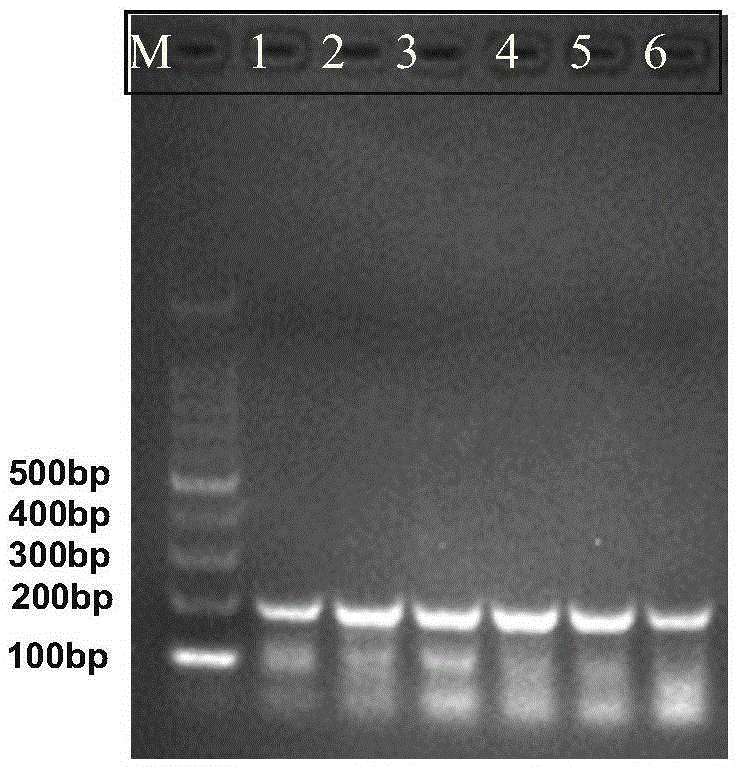
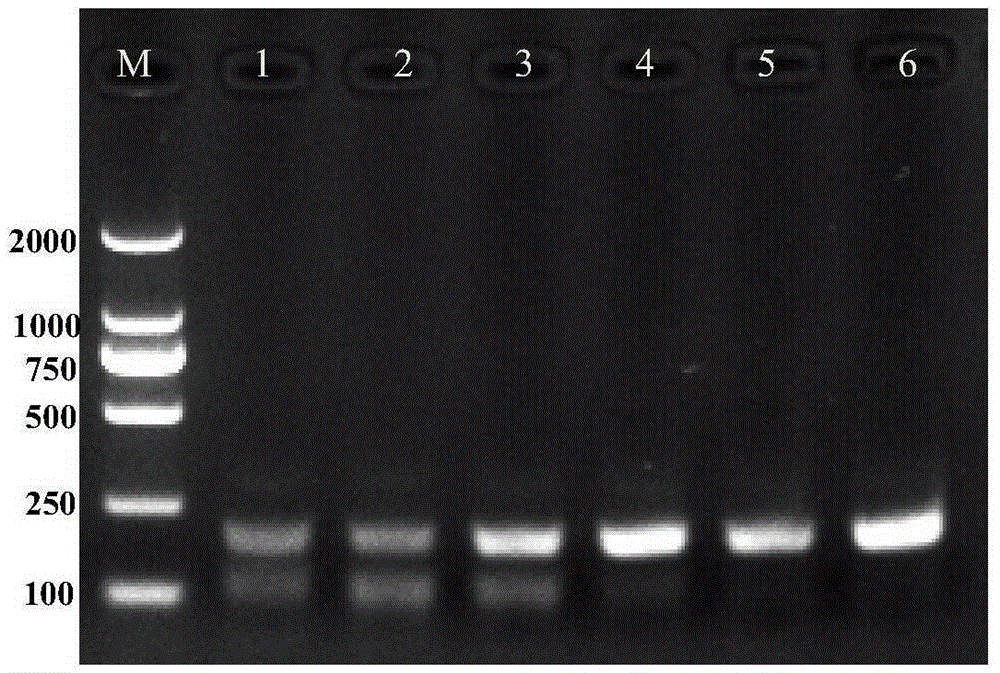
[0072] Embodiment 3, the sensitivity analysis of RPA detection
[0073] The total DNA of the quarantine Solanum weed Solanum aureus was diluted from 20.8ng / μL to 5.2, 1.04, 0.52, 0.104, 0.052, 0.0104ng / μL for RPA detection. In a 25 μL reaction system, 1 μL of the above solution was added to obtain 5.2, 1.04, 0.52, 0.104, 0.052, and 0.0104 ng of DNA templates. Refer to Step 2 of Example 1 for other operations.
[0074] The results of the RPA reaction showed that when the DNA template was 0.0104ng, the target fragment (210bp, sequence 3) was relatively clear, which proved that the RPA reaction had higher sensitivity. Experimental results such as image 3 shown.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com