Fusion protein molecular weight analysis method
A technology of fusion protein and analysis method, applied in the direction of biological testing, material inspection products, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0033] Example 1: Analysis of the complete molecular weight of the fusion protein (removal of N-sugar first, and then removal of sialic acid)
[0034] Fusion protein complete molecular weight analysis steps:
[0035] (1) Removal of N-Glycosylations
[0036] Get 50 μ g Etanercept fusion protein sample (Pfizer Pharmaceutical Co., Ltd., the same below), add 50 mM NH 4 HCO 3 (pH8.0) solution to a volume of 20 μL, add 1 μL of 4-fold diluted peptide N-glycosidase PNGaseF (NEB, 500000u / mL), mix well, and incubate at 37°C for 24 hours.
[0037] (2) Sialic acid removal
[0038] For the sample incubated in (1), adjust the pH value to 5-6 with 1% FA, add 0.5 μL of sialidase Neuraminidase (Roche, 1 U / 100 μL), and incubate at 37° C. for 24 hours.
[0039] (3) ESI-Q-TOFMS analysis
[0040] The fusion protein of asialo de-N-glycans adopts 50mM NH 4 HCO 3 (pH8.0) solution was diluted to 1 mg / mL; then MassPREP™ MicroDesaltingColumn 2.1 × 5mm desalting column (Waters, the same below) aft...
Embodiment 2
[0042] Example 2: Analysis of the complete molecular weight of the fusion protein (sialic acid is removed first, and then N-sugar is removed)
[0043] Fusion protein complete molecular weight analysis steps:
[0044] (1) Sialic acid removal
[0045] Take 50μg Etanercept fusion protein sample, add 50mM NH 4 HCO 3 (pH 8.0) solution to a volume of 20 μL, adjusted the pH value to 5-6 with 1% FA, added 0.5 μL of sialidase Neuraminidase, and incubated at 37° C. for 24 hours.
[0046] (2) N-sugar removal
[0047] The sample incubated in (1), with 1% NH 3 .H 2 O to adjust the pH value to 7-8, add 1 μL of 4-fold diluted PNGaseF (NEB, 500000u / mL), and incubate at 37°C for 24 hours.
[0048] (3) ESI-Q-TOFMS analysis
[0049] De-N-sugar asialo fusion protein using 50mM NH 4 HCO 3 (pH8.0) solution was diluted to 1mg / mL; then MassPREPTMMicroDesaltingColumn2.1 × 5mm desalting column desalting, and finally mass spectrometry analysis.
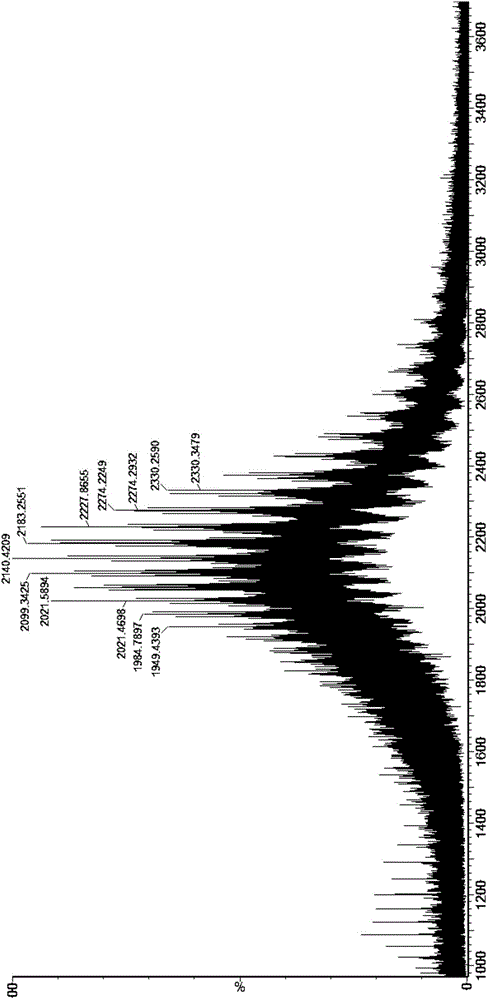
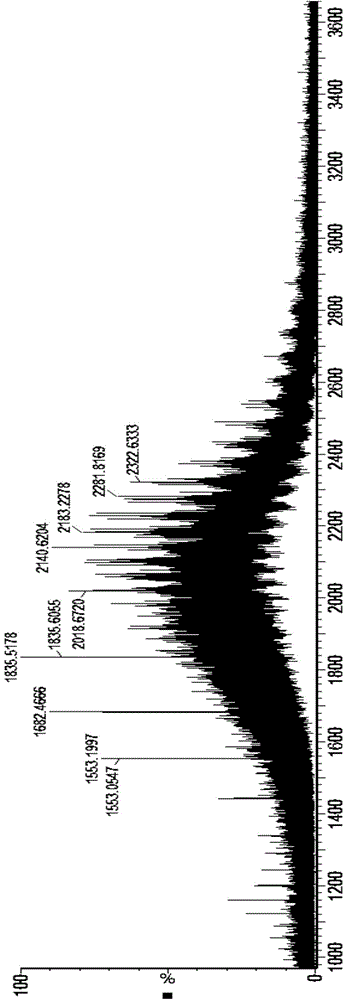
[0050] Experimental results: See the attachment...
Embodiment 3
[0051] Example 3: Comparison of molecular weight analysis of asialo deconvoluted intact protein under different pH conditions after N-sugar removal
[0052] Fusion protein complete molecular weight analysis steps:
[0053] (1) N-sugar removal
[0054] Take 400μg Etanercept fusion protein sample, add 50mM NH 4 HCO 3 (pH8.0) solution to a volume of 160 μL, add 2 μL PNGaseF, mix well, and incubate at 37°C for 24 hours.
[0055] (2) Sialic acid removal
[0056] Take 4 samples incubated in (1), 50 μg each, adjust the pH to 4, 5, 6, and 7 with 1% FA, add 0.5 μL of sialidase Neuraminidase, and incubate at 37°C for 24 hours.
[0057] (3) ESI-Q-TOFMS analysis
[0058] The fusion protein of de-N-sugar and asialic acid uses 50mM NH 4 HCO 3 (pH8.0) solution diluted to 1mg / mL; then MassPREPTMMicroDesaltingColumn2.1 × 5mm after desalting, mass spectrometry.
[0059] Experimental results: After N-sugar removal, the molecular weight comparison analysis results of asialic acid deconvol...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com