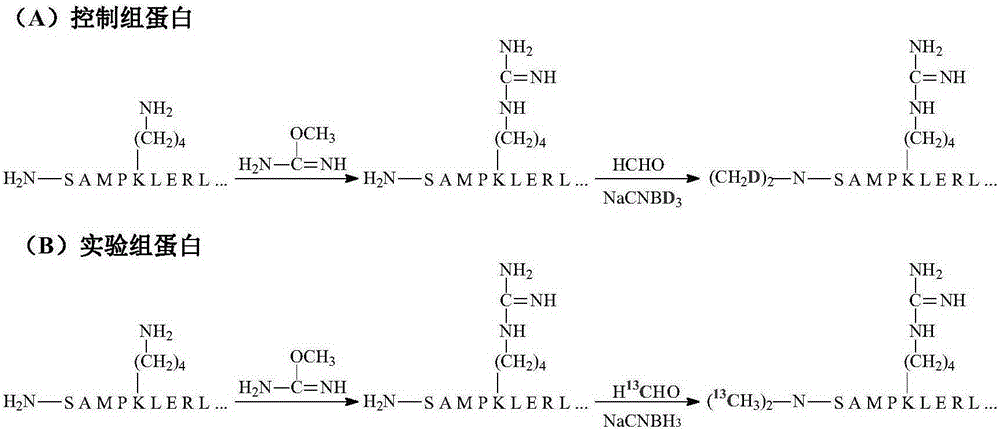Patents
Literature
84 results about "Intact protein" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Intact Protein Characterization. Full characterization of a protein by mass spectrometry includes determination of the protein sequence, and identification and relative quantitation of protein isoforms, including identification and localization of one or multiple post-translational modifications (PTMs).
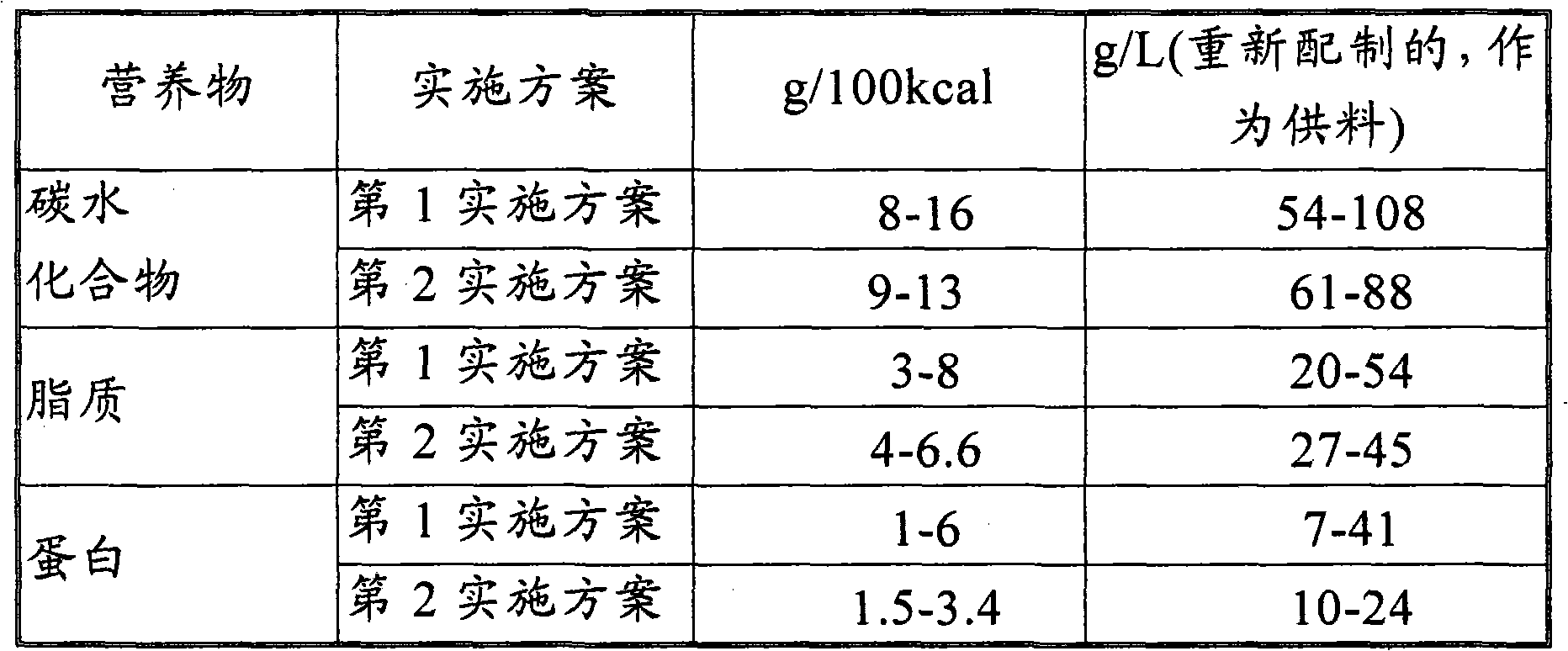
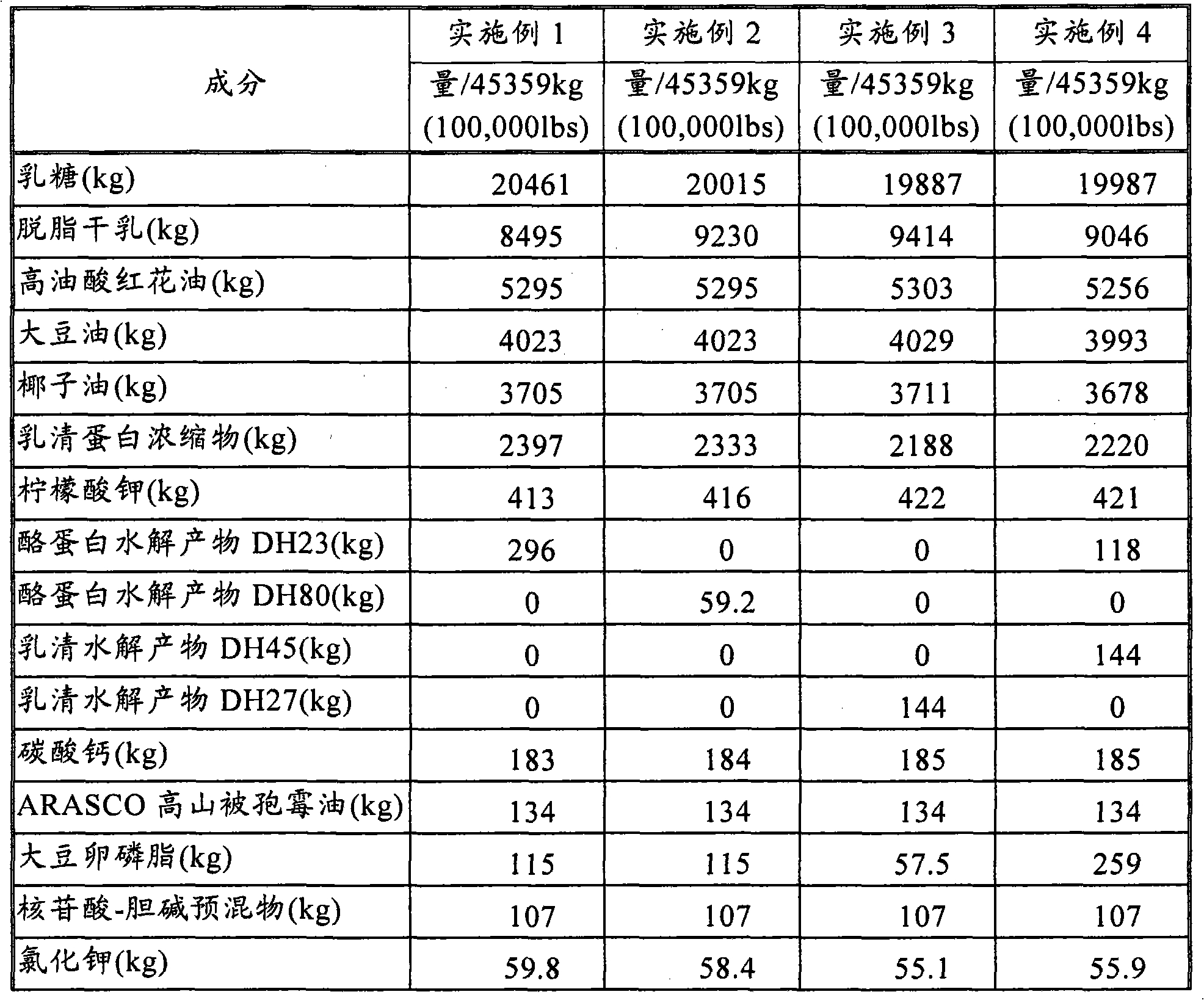
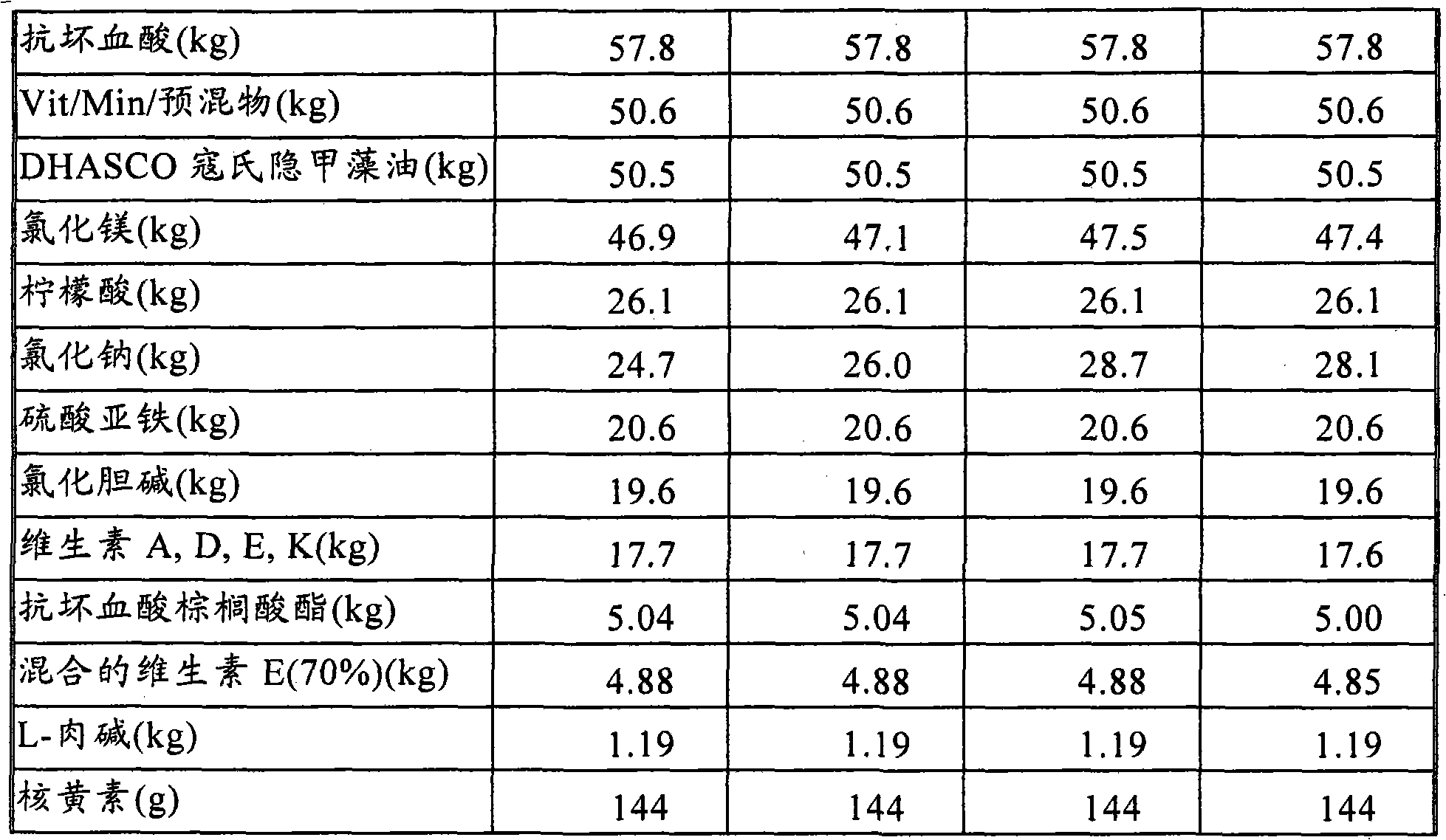
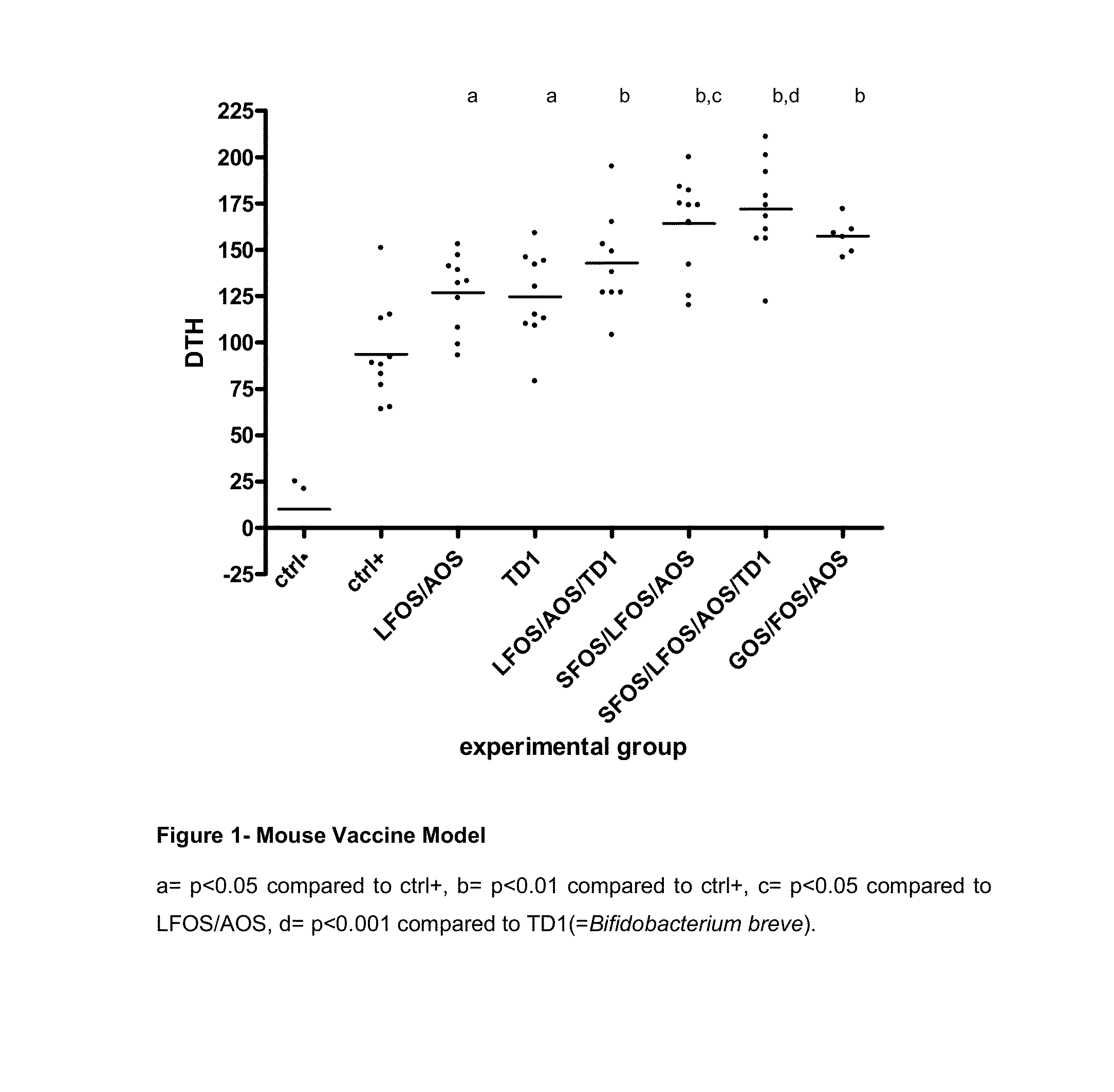
Nutritional composition intended for specific gastro-intestinal maturation in premature mammals
InactiveUS6733770B1Good nutritional needsMinimize nitrogen wasteBiocidePeptide/protein ingredientsComplete proteinIntact protein
A nutritional enteral composition intended for favoring the growth and maturation of non-mature gastro-intestinal tracts of young mammals, which contains as a protein source a mixture of dietary protein hydrolysates and intact proteins being partly in the form of bioactive peptides.
Owner:NESTEC SA
Method for kidney disease detection
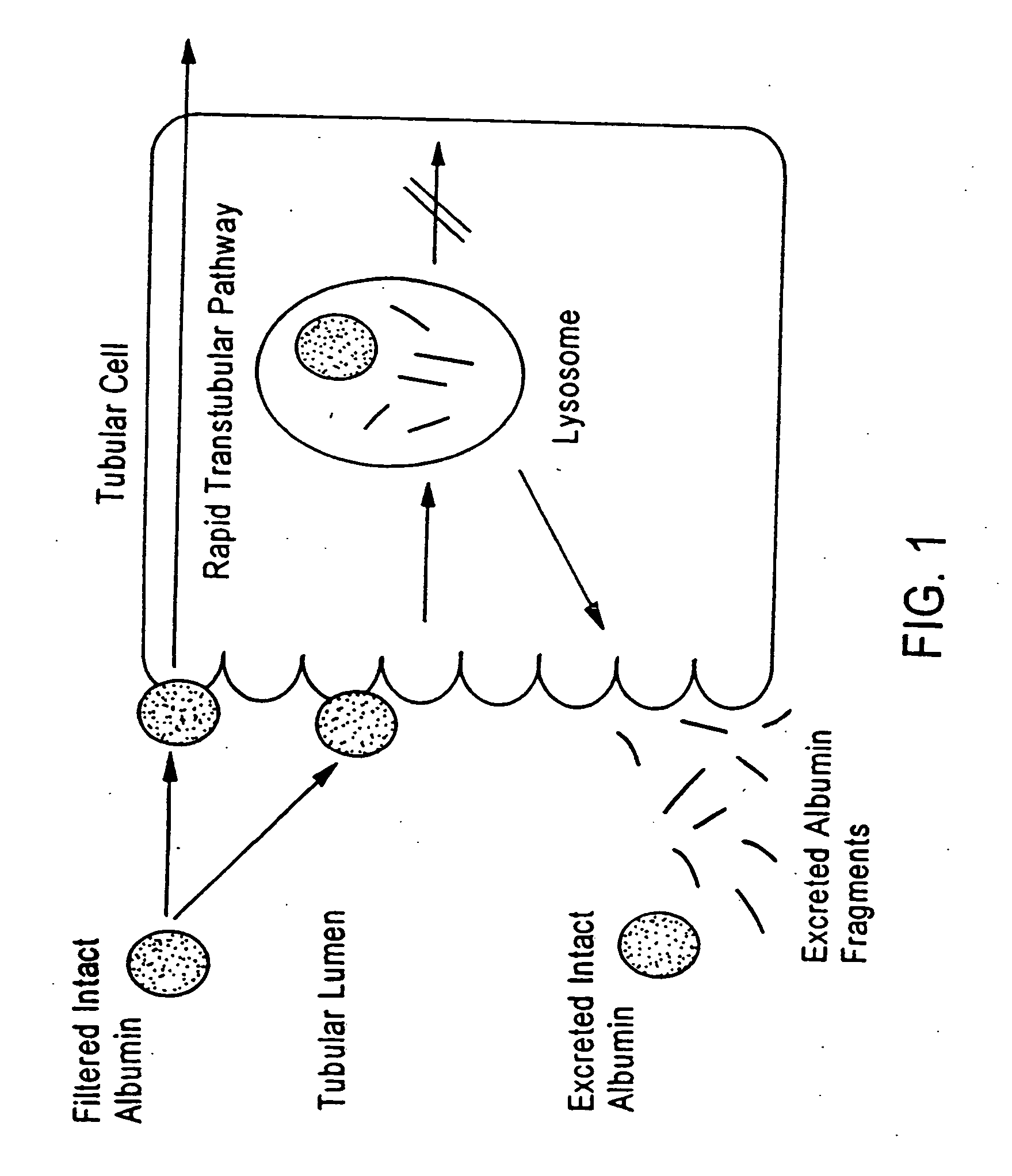
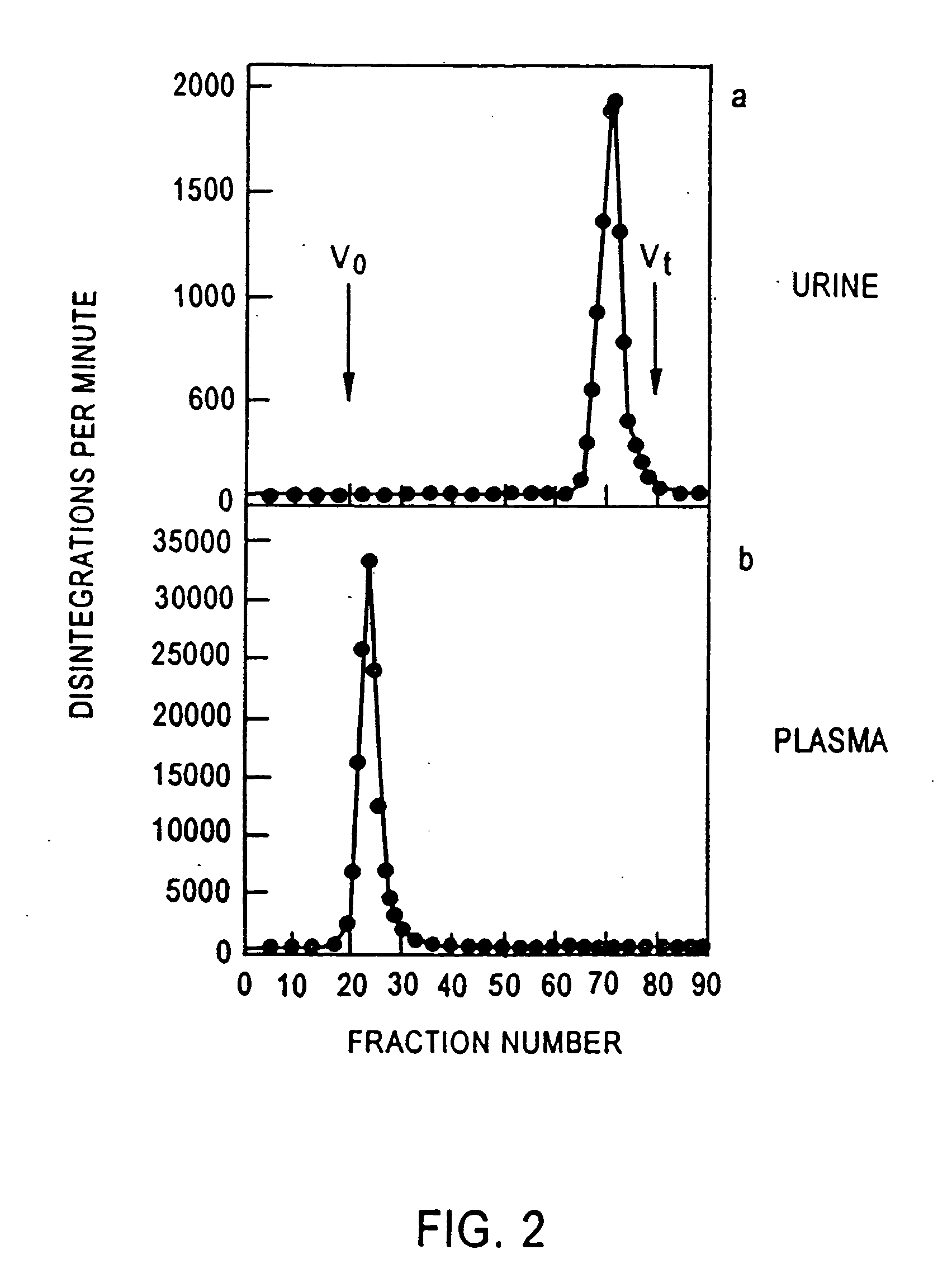
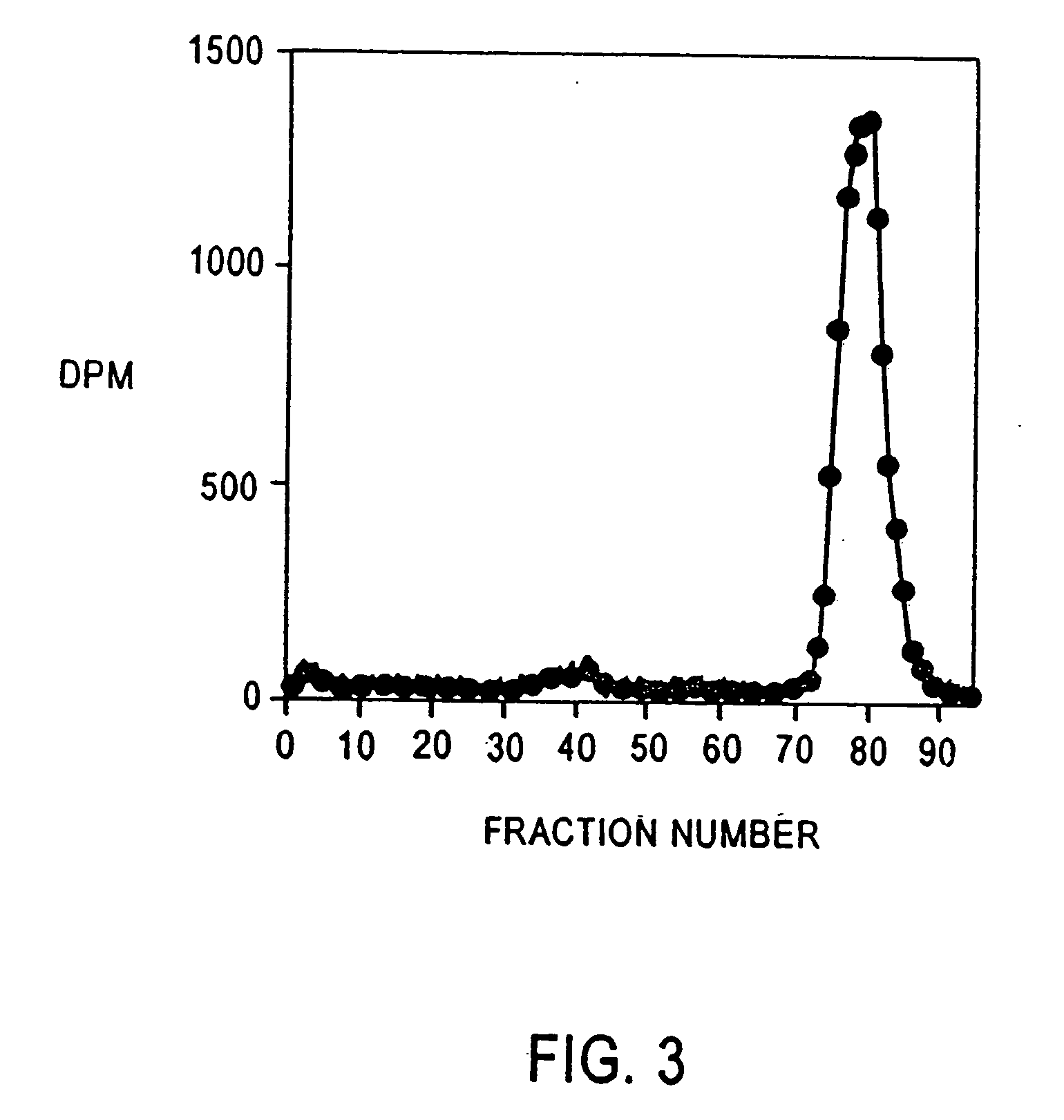
A method for diagnosing early stage renal disease and / or renal complications of a disease in which intact albumin is an indicator of the renal disease and / or complications. The method includes an isolated intact protein, an anti-intact protein antibody thereto, and methods for preparing the same.
Owner:MONASH UNIV
Shelf stable nutritional composition containing intact whey protein, process of manufacture and use
ActiveUS7247320B2High protein concentrationIncrease concentrationMammal material medical ingredientsWhey manufactureHigh concentrationIntact protein
A composition is provided that provides a nutritionally complete calorically dense formula suitable for use as a ready-to-use liquid composition that does not require reconstitution and admixing, which contains intact whey protein in high concentration and which is shelf stable for up to 6 months or more at ambient temperature.
Owner:SOC DES PROD NESTLE SA
Stable nutritional powder
ActiveCN101951789AImprove oxidation stabilityEnhance sensory effectFood preparationIntact proteinWhey protein
Disclosed are compositions comprising carbohydrate; lipid, comprising from about 0.25% to about 2.5 % lecithin by weight of total lipid; from about 90% to about 99.5% of intact protein by weight of total protein; and from about 0.5% to about 10% of at least one hydrolyzed protein selected from the group consisting of hydrolyzed casein protein and hydrolyzed whey protein; wherein the hydrolyzed protein has a degree of hydrolysis of between about 23% and about 90%, and wherein the compositions are nutritional powders. The nutritional powders provide improved oxidative stability and sensory performance.
Owner:ABBOTT LAB INC
Nutrition bar
A nutrition or other food bar which includes, preferably in moderate to high levels, peptides in the form of hydrated high water activity peptides. The peptides are typically either intact proteins or hydrolyzed proteins. Inclusion of peptides in the form of high water activity peptides helps prevent migration of water from sugars to the proteins which would otherwise result in formation of hard crystalline sugars and food bars which are excessively hard. Whey protein isolate and hydrolyzed whey protein are preferred high water activity peptides.
Owner:SLIM FAST FOODS
Nutritional Compositions Containing A Peptide Component with Anti-Inflammatory Properties and Uses Thereof
ActiveUS20140271553A1Reducing proinflammatory responseReduce productionBiocideAntipyreticIntact proteinInterleukin 17
The present disclosure relates to nutritional compositions comprising a protein equivalent source, wherein 20% to 80% of the protein equivalent source includes a peptide component comprising SEQ ID NO 4, SEQ ID NO 13, SEQ ID NO 17, SEQ ID NO 21, SEQ ID NO 24, SEQ ID NO 30, SEQ ID NO 31, SEQ ID NO 32, SEQ ID NO 51, SEQ ID NO 57, SEQ ID NO 60, and SEQ ID NO 63, and 20% to 80% of the protein equivalent source comprises an intact protein, a partially hydrolyzed protein, or combinations thereof. The disclosure further relates to methods of reducing the inflammatory response and / or production of proinflammatory cytokines, i.e. Interleukin-17, by providing said nutritional compositions to a target subject.
Owner:MEAD JOHNSON NUTRITION
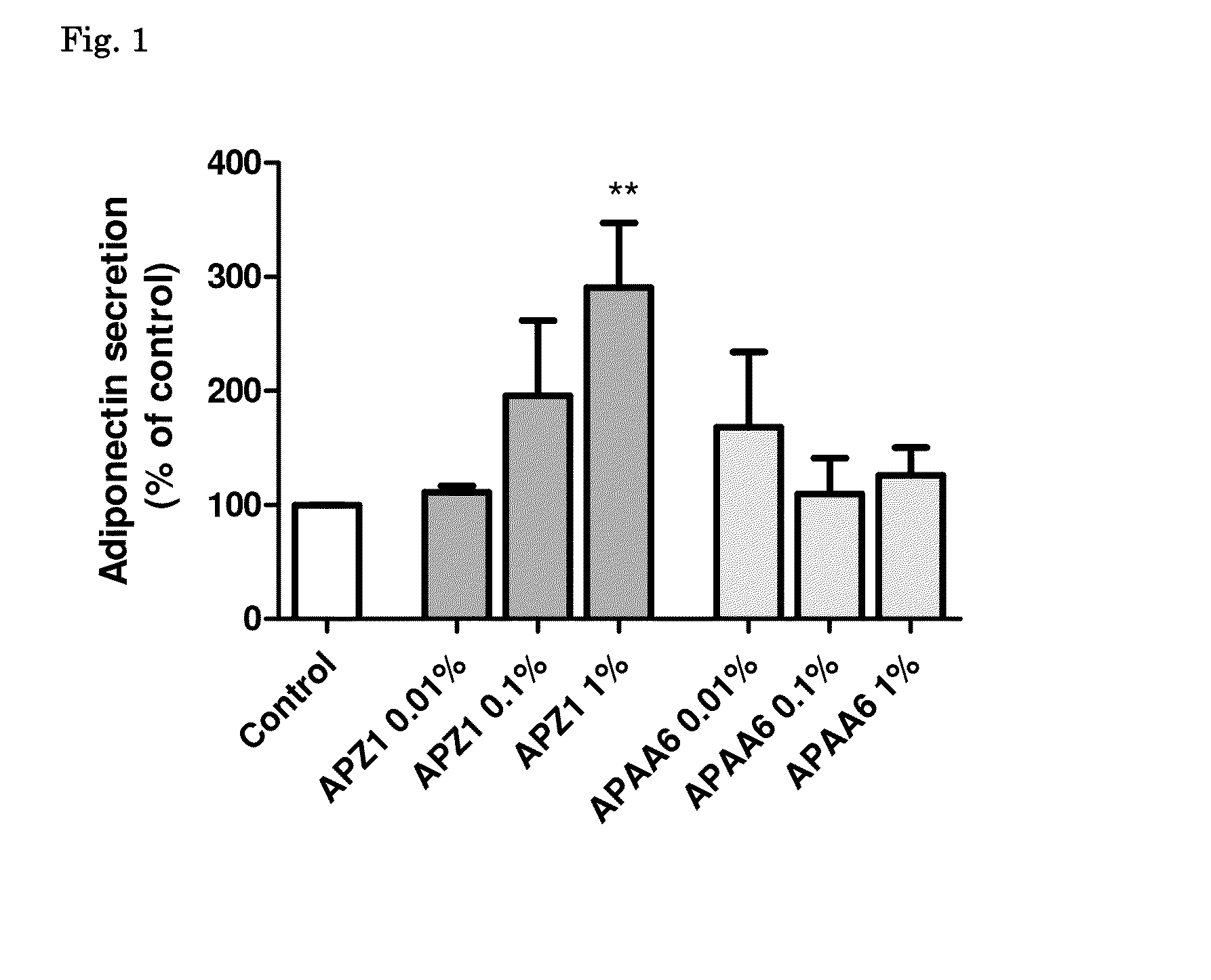
Nutritional Composition Containing a Peptide Component with Adiponectin Simulating Properties and Uses Thereof
ActiveUS20140271586A1Body weightSatisfies requirementBiocideTetrapeptide ingredientsComplete proteinIntact protein
The present disclosure relates to nutritional compositions including a protein equivalent source that includes a peptide component comprising selected peptides. The protein equivalent source may further include intact protein, hydrolyzed protein, including partially hydrolyzed protein, or combinations thereof. The disclosure further relates to methods of promoting healthy body weight in a target subject by stimulating adiponectin levels by providing the nutritional compositions disclosed herein to a target subject, which includes a pediatric subject.
Owner:MEAD JOHNSON NUTRITION
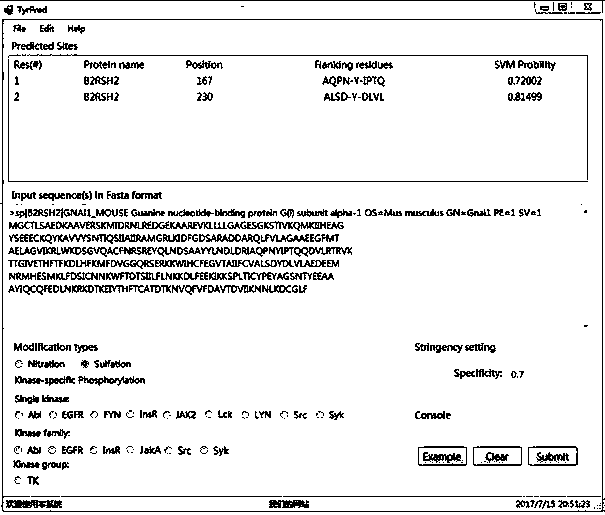
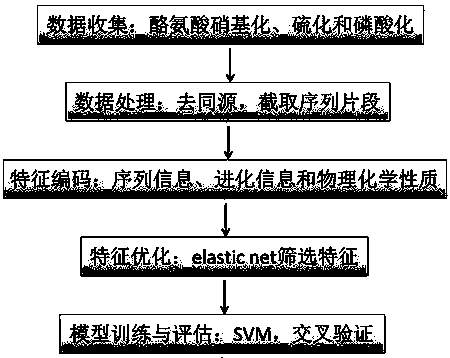
Prediction algorithm for recognizing tyrosine posttranslational modification sites
InactiveCN107463795AImprove predictive performanceImprove forecast qualityBiostatisticsProteomicsIntact proteinPrediction algorithms
The invention discloses a prediction algorithm for recognizing tyrosine posttranslational modification sites. The algorithm comprises the steps of data collection, data processing, feature coding, feature optimization and model training and evaluation. The invention furthermore discloses application of the prediction algorithm. According to the algorithm, features of the tyrosine posttranslational modification sites are extracted comprehensively from the perspectives of protein sequence information, evolutional information and physical and chemical properties, Elastic Net is used as an optimization means to automatically select variables to screen multidimensional features, redundant information is removed, a prediction model of tyrosine nitration, sulfuration and phosphorylation sites is constructed in combination with an SVM, the prediction capability of the prediction model is improved, and the prediction quality of the tyrosine posttranslational modification sites is remarkably improved. Through a developed prediction software platform TyrPred, predictive analysis of nitration modification sites, sulfuration modification sites and phosphorylation modification sites of tyrosine on intact protein is realized, and a convenient, economical and rapid research tool and important reference are provided for research of tyrosine posttranslational modification.
Owner:NANCHANG UNIV
Method of treating or preventing obeisity and lipid metabolism disorders and compositions for use therein
InactiveUS20060105938A1Lose weightUndesired impactBiocideHydrolysed protein ingredientsComplete proteinIntact protein
The present invention provides a method of preventing or treating human obesity, said method comprising ingesting a composition containing, calculated on dry matter: 10-100 wt. % protein hydrolysate; 0-90 wt. % intact protein; 0-50 wt. % carbohydrate; and wherein hydrolysed protein and intact protein together are present in a concentration (w / w) that exceeds the carbohydrate concentration (w / w). The invention also encompasses the use of the same composition in a method of preventing or treating lipid metabolism disorders and in a method for improving body appearance. Other aspects of the invention relate to nutritional beverages, snacks and soups that can advantageously be employed in accordance with the aforementioned methods.
Owner:KERRY GROUP SERVICES
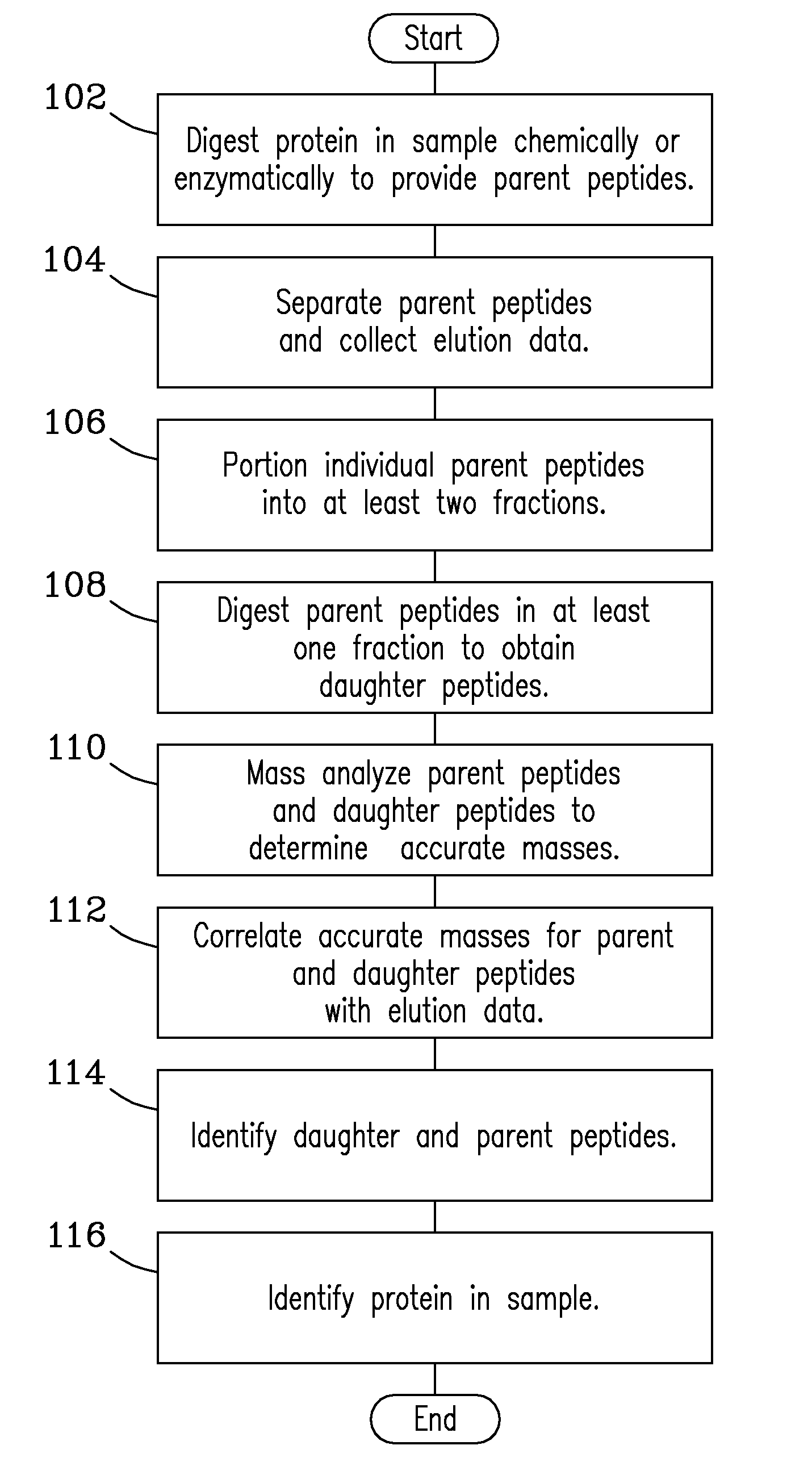
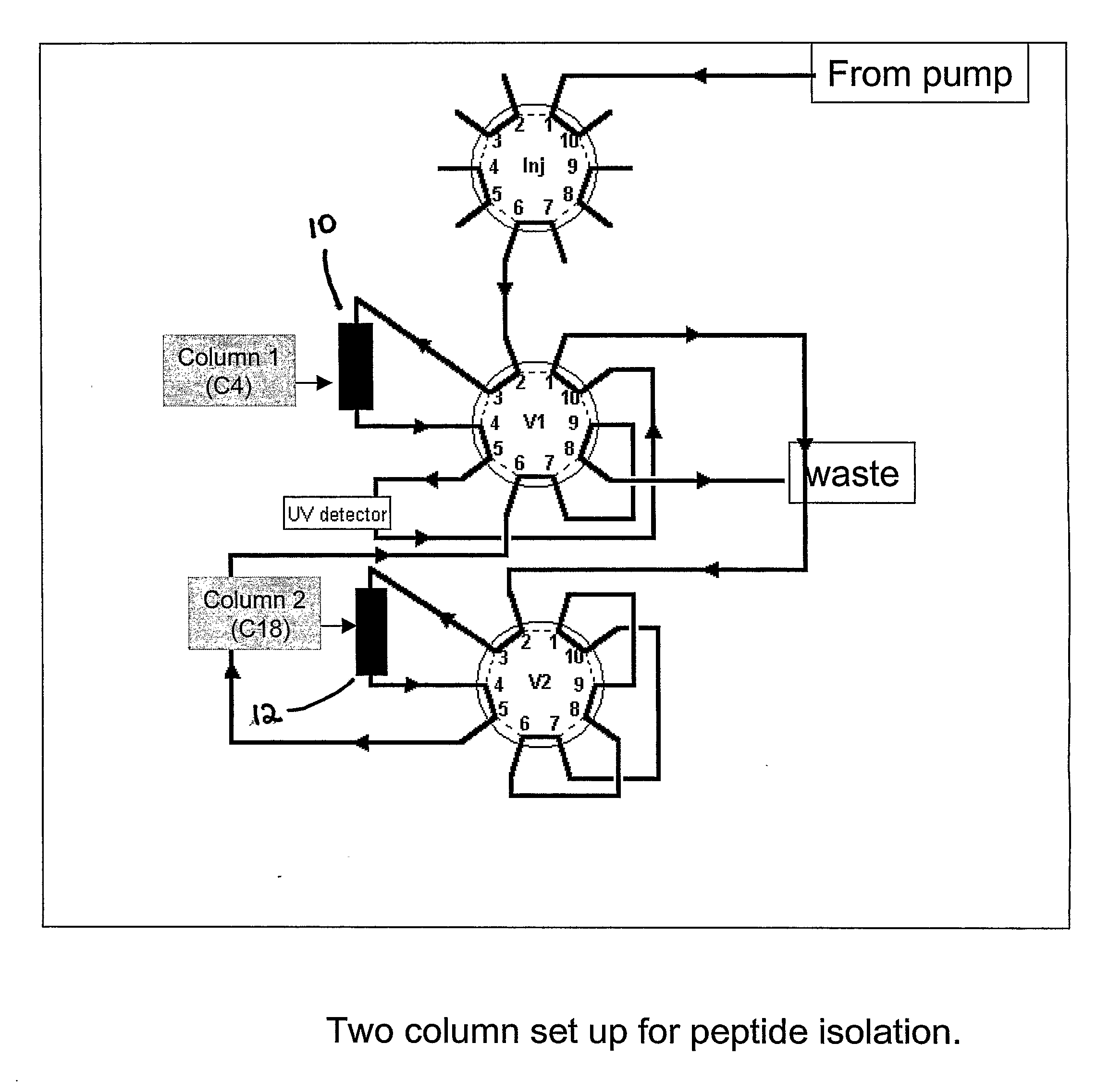
Solution fragmentation systems and processes for proteomics analysis
InactiveUS20090256068A1Accurate identificationEasy to understandParticle separator tubesPeptide/protein ingredientsComplete proteinIntact protein
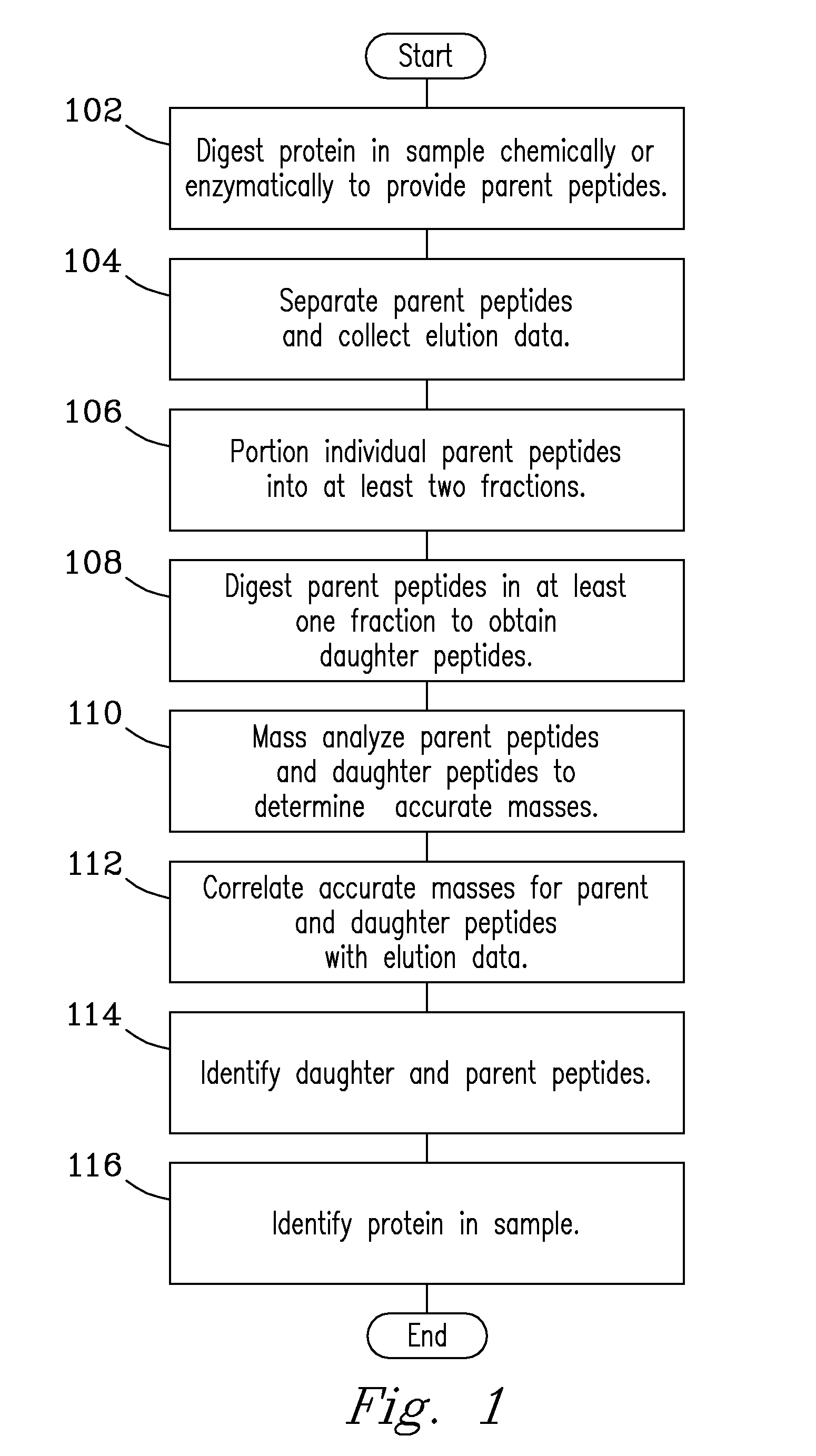
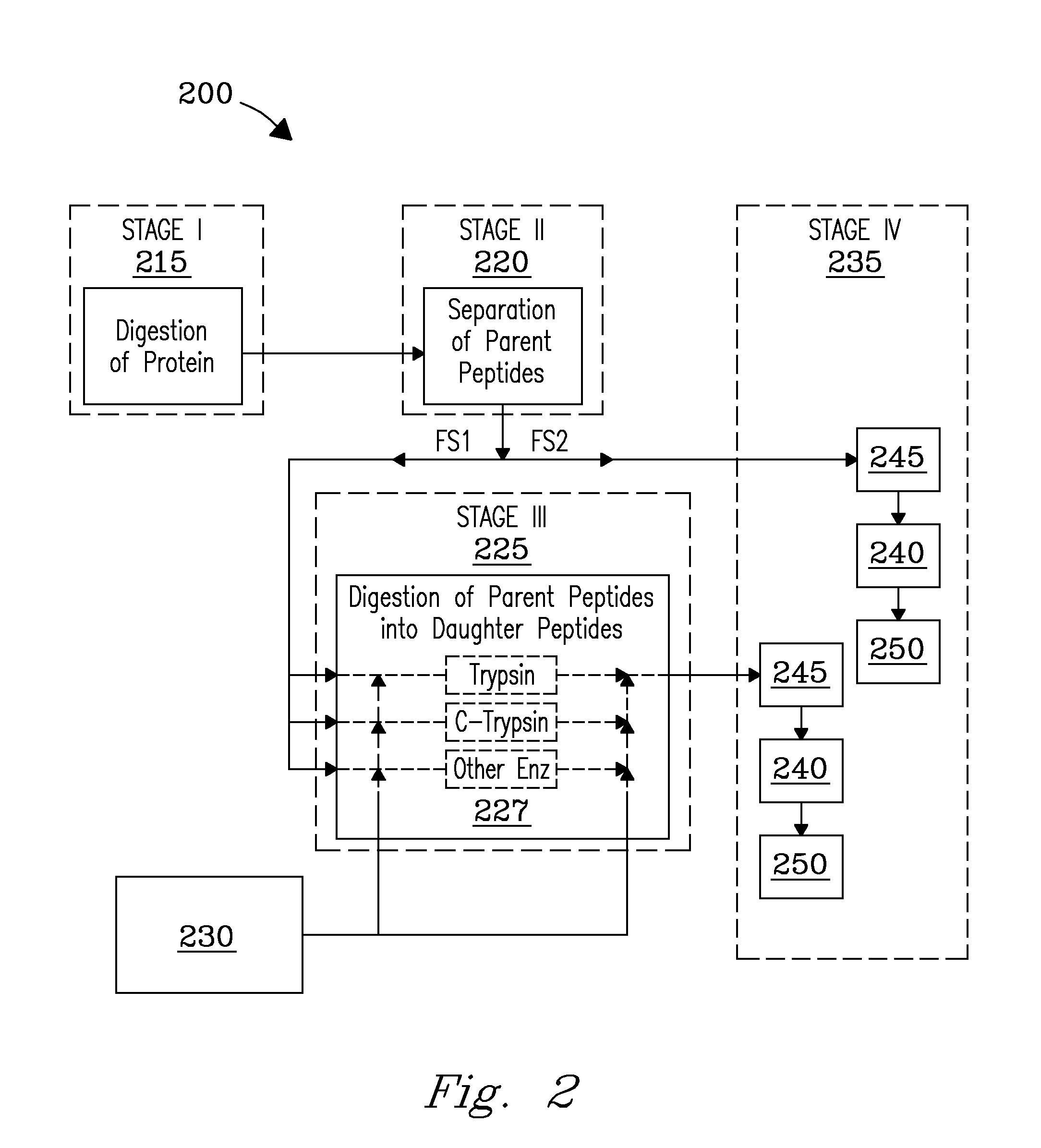
A solution-phase digestion process is described. Intact proteins are digested to obtain parent peptides, which are separated and subsequently mass analyzed. Individual parent peptides are digested to obtain daughter peptides, which are also subsequently mass analyzed. Accurate mass data obtained from mass analysis of both parent and daughter peptides are correlated with separations data obtained during separation of the parent peptides to provide peptide identification. The process is expected to provide unique peptides by which to identify intact proteins in a sample without need for MS / MS gas-phase fragmentation.
Owner:BATTELLE MEMORIAL INST
Protein free formula
ActiveUS20100172876A1Avoid developmentReduce severityBiocidePeptide/protein ingredientsIntact proteinFeces
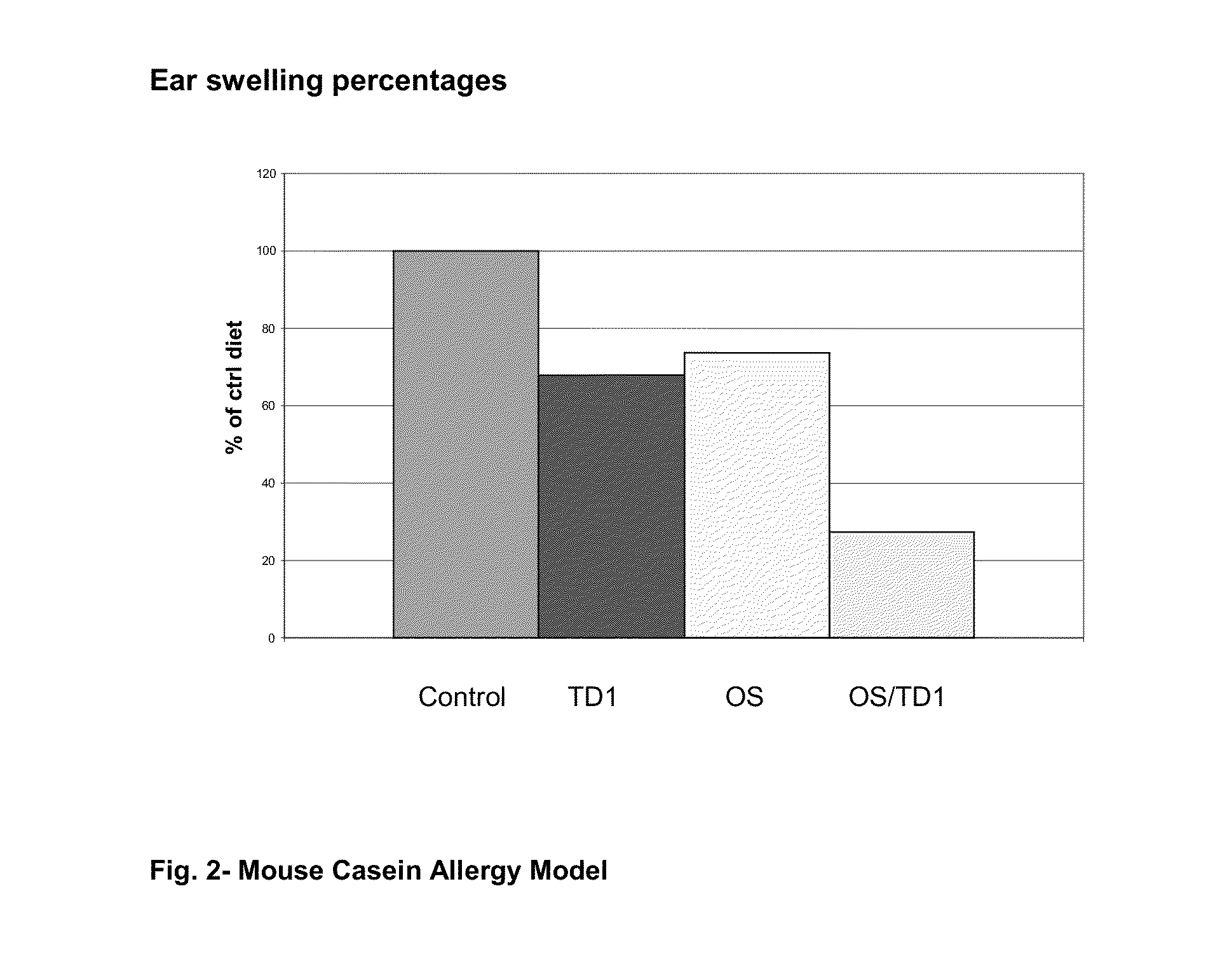
The invention relates to the use of composition comprising free amino acids as a sole source of protein, a fatty acid source comprising long chain polyunsaturated fatty acids, a carbohydrate source comprising digestible and indigestible carbohydrates, and milk protein free Bifidobacteria for treating a person suffering from (a) colic, congestion, runny nose, wheezing, vomiting, diarrhoea, bloody stools, mucus in stools, rash, eczema, gastroesophageal reflux, eosinophilic esophagitis or asthma; (b) cow's milk allergy and / or food protein intolerance; and / or (c) infections, wherein the indigestible carbohydrate is selected from a milk protein free source and the total composition is essentially free of intact proteins.
Owner:SHS INTERNATIONAL
Protein free formula
ActiveUS8691213B2Avoid developmentReduce severityBiocidePeptide/protein ingredientsSerum rashIntact protein
The invention relates to the use of composition comprising free amino acids as a sole source of protein, a fatty acid source comprising long chain polyunsaturated fatty acids, a carbohydrate source comprising digestible and indigestible carbohydrates, and milk protein free Bifidobacteria for treating a person suffering from (a) colic, congestion, runny nose, wheezing, vomiting, diarrhea, bloody stools, mucus in stools, rash, eczema, gastroesophageal reflux, eosinophilic esophagitis or asthma; (b) cow's milk allergy and / or food protein intolerance; and / or (c) infections, wherein the indigestible carbohydrate is selected from a milk protein free source and the total composition is essentially free of intact proteins.
Owner:SHS INTERNATIONAL
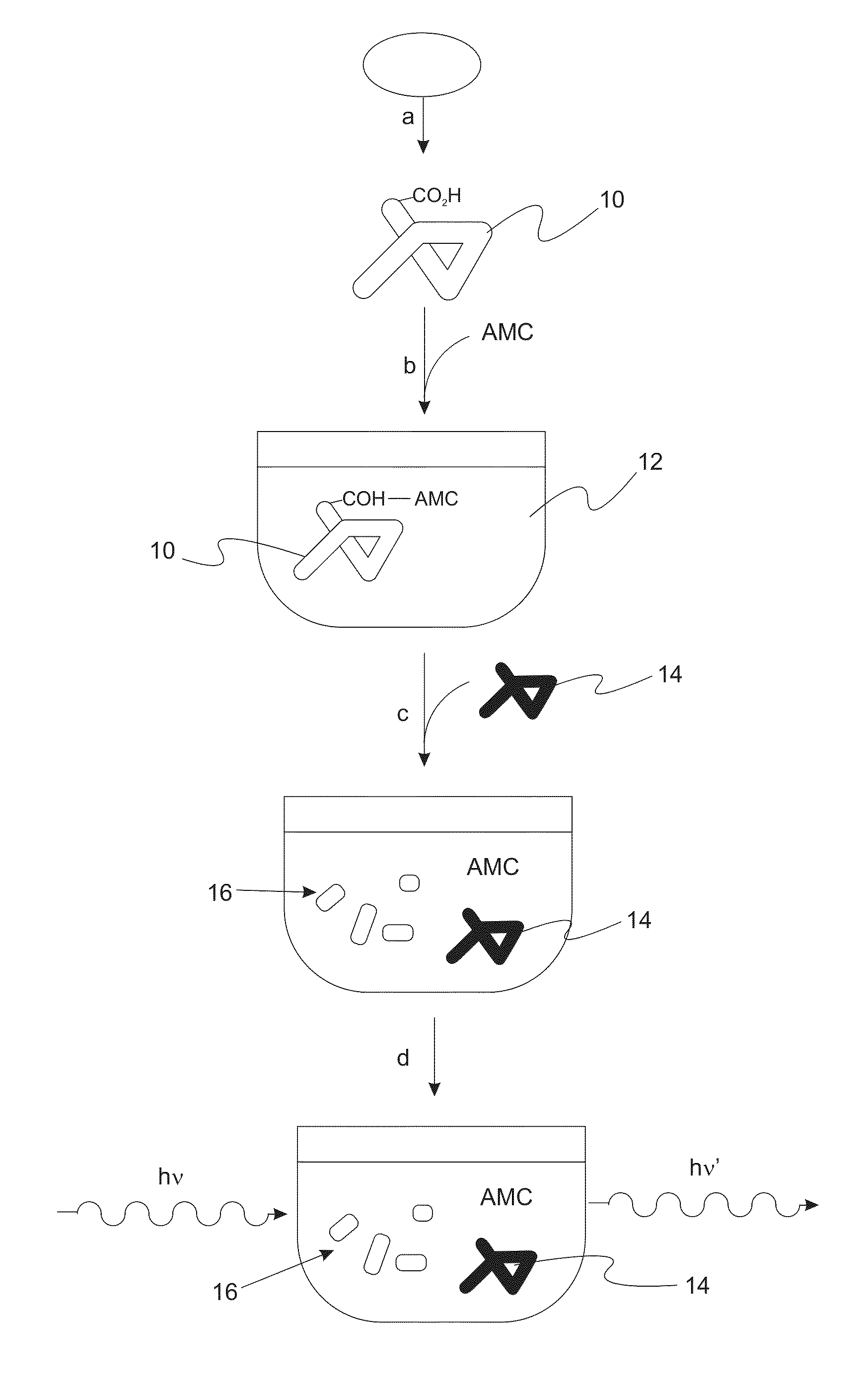
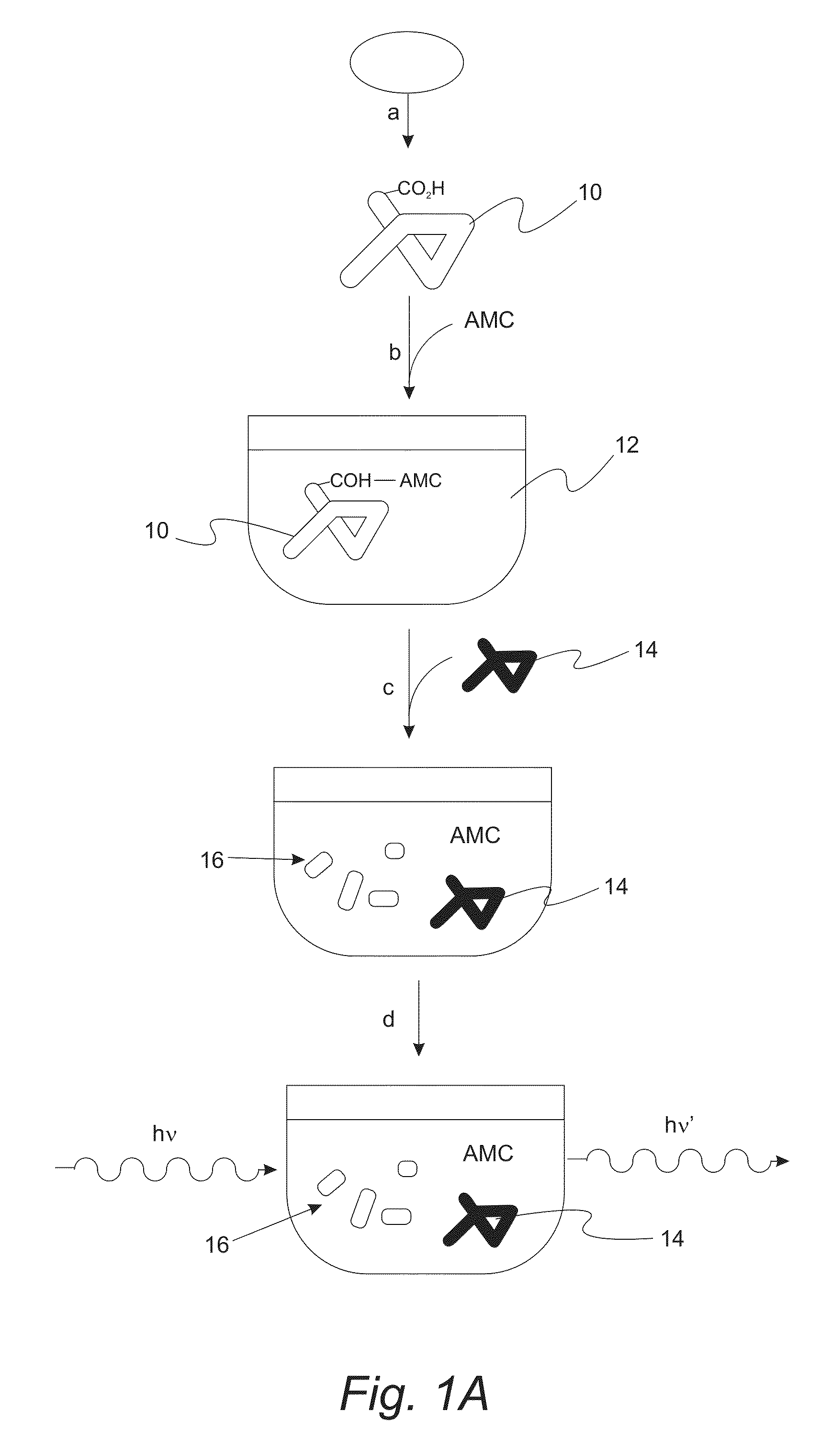
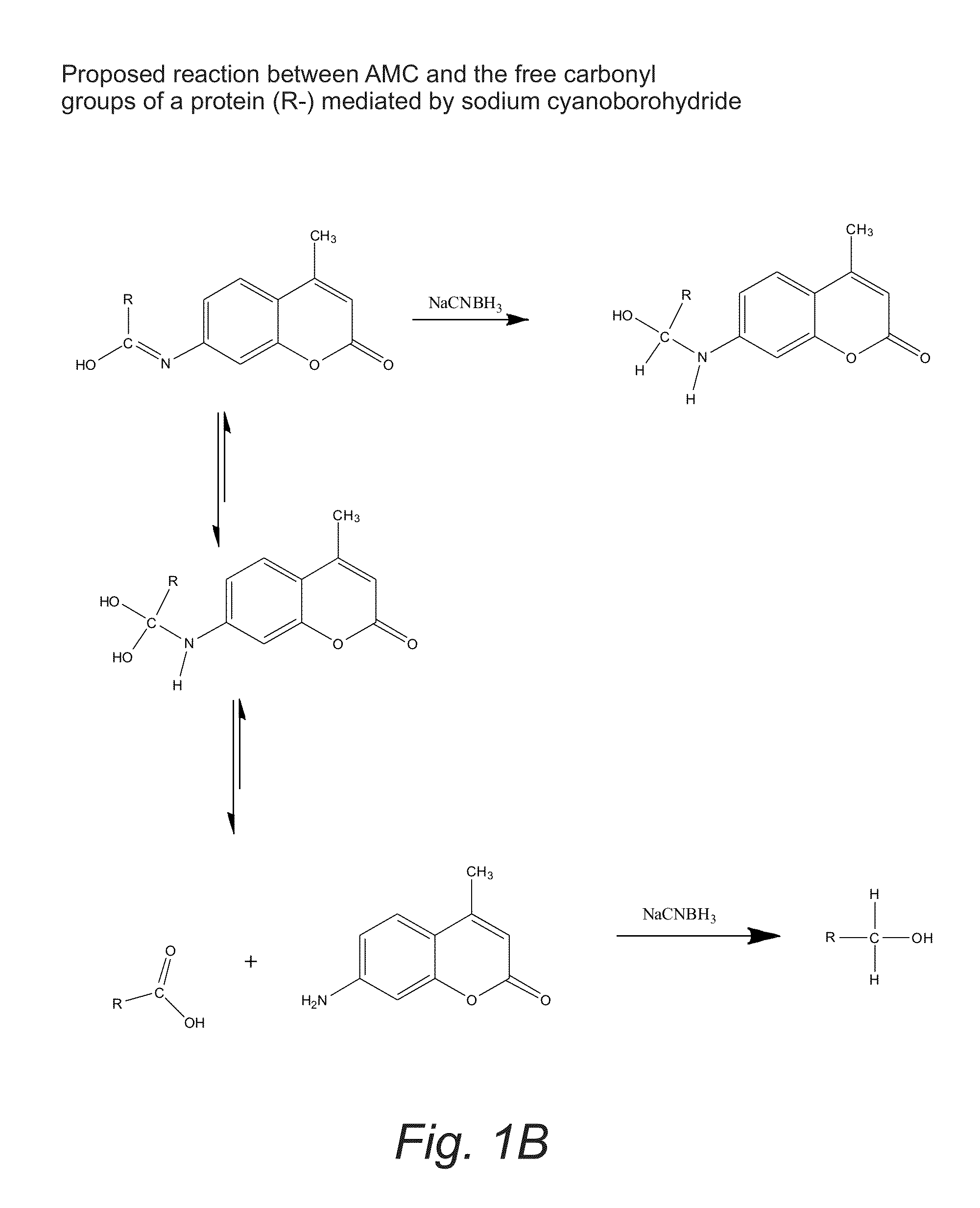
Labeling of Proteins with the Fluorophore 7-amino-4-methylcoumarin (AMC) Generated Novel Proteolytic Substrates
ActiveUS20130059321A1Stable to oxidationIncreased proteolytic susceptibilityMicrobiological testing/measurementFluorescence/phosphorescenceIntact protein7-amino-4-methylcoumarin
A method of measuring the degradation of intact proteins includes a step of providing a protein substrate having one or more free or exposed carboxyl groups and then reductively attaching 7-amino-4-methylcoumarin (AMC) to the protein substrate with a reducing agent. The protein substrate is contacted in a test solution with one or more proteolytic enzymes that degrade the protein substrate. The amount of AMC attached to the protein substrate is then determined by monitoring the fluorescence of free 7-amino-4-methylcoumarin that is formed during degradation of the protein substrate to protein fragments.
Owner:UNIV OF SOUTHERN CALIFORNIA
Detection of Disease Associated Proteolysis
InactiveUS20090035797A1Inherent problemParticle separator tubesPeptide/protein ingredientsIntact proteinOrganism
Described herein are methods and techniques to study the “degradome”. The degradome of a specific protease is the complete product of the natural substrate repertoire of that enzyme in a cell, tissue or organism. The complete set of proteases that are expressed at a particular moment or circumstance by a cell, tissue or organism produces the collective degradome. Included in the methods described herein are approaches that allow the direct identification and characterization of degradome peptides from approx. 400 to approx. 12,000 Da. The methods of the invention avoid the inherent problems of studying the peptidome by focusing on specific or unique proteolytic cleavages that occur as a result of endogenous protease activity induced by specific diseases. Once characterized, the presence of, or change in level of, specific peptides of the degradome can be used, e.g., to identify specific peptides having elevated levels compared to a reference normal / or to correlate identified peptides with specific proteins and / or to identify protein fragmentation patterns (e.g., peptide ladders) and the specific protease(s) that brought them about and then correlate this information with the presence or absense of a specific disease or condition. Thus, the methods of the invention can be used, for example, to identify new diagnostic markers and / or therapeutic targets, as specific clinical diagnostic methods for individual patients and as methods of monitoring the progress of a therapeutic regimen for the treatment of a patient.
Owner:NORTHEASTERN UNIV
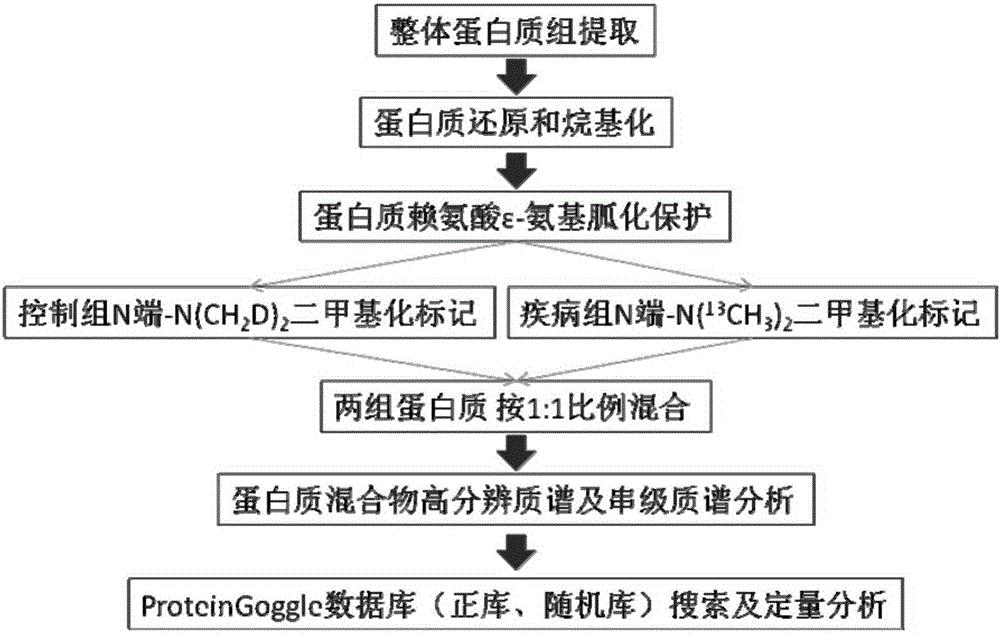
Quantitative analysis method for intact protein under different physiological or pathological conditions
InactiveCN105242050AImprove labeling efficiencyImprove accuracyBiological testingData setIntact protein
The invention relates to a quantitative analysis method for intact protein under different physiological or pathological conditions. The method comprises firstly taking two groups of intact protein under different physiological or pathological conditions respectively as a control group and a disease group, and performing reduction and alkylation; performing chemical protection on epsilon-amino on lysine residue in the two groups of the alkylated protein sequences; performing isobaric marking on the N-end amino of the two groups of protein subjected to chemical protection; mixing the two groups of protein subjected to isobaric marking according to equal proportions, and performing high-resolution mass spectrometry and tandem mass spectrometry analysis to obtain a data set; and performing qualitative and quantitative database search on the data set, so as to obtain protein ID and the relative proportion of each protein relative to the protein in the control group, in other words, the up-regulation or down-regulation situations of all protein under disease conditions. Compared with the prior art, the method is high in marking efficiency and high in accuracy, and is suitable for quantitative analysis on intact protein based on high-resolution tandem mass spectrometry.
Owner:TONGJI UNIV
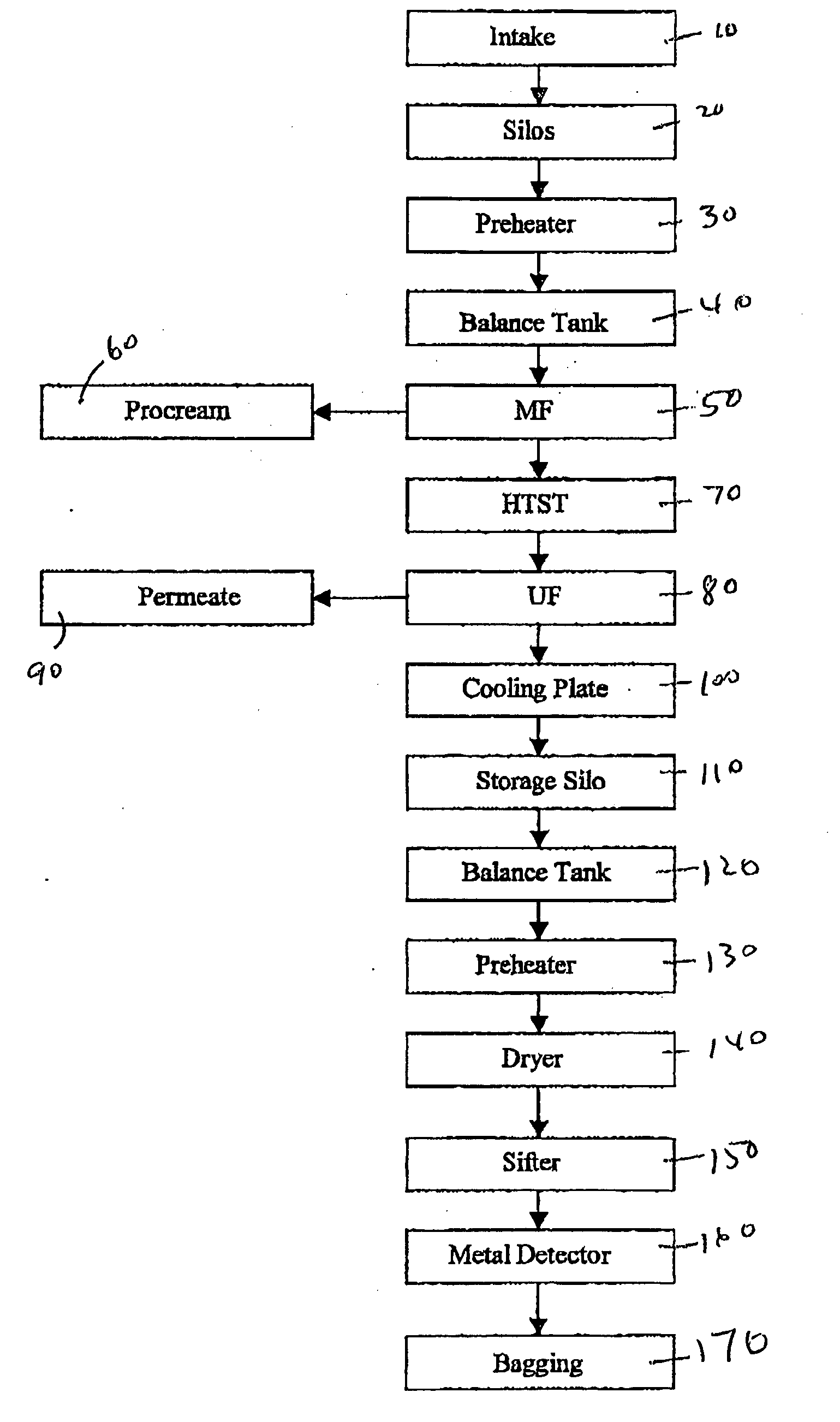
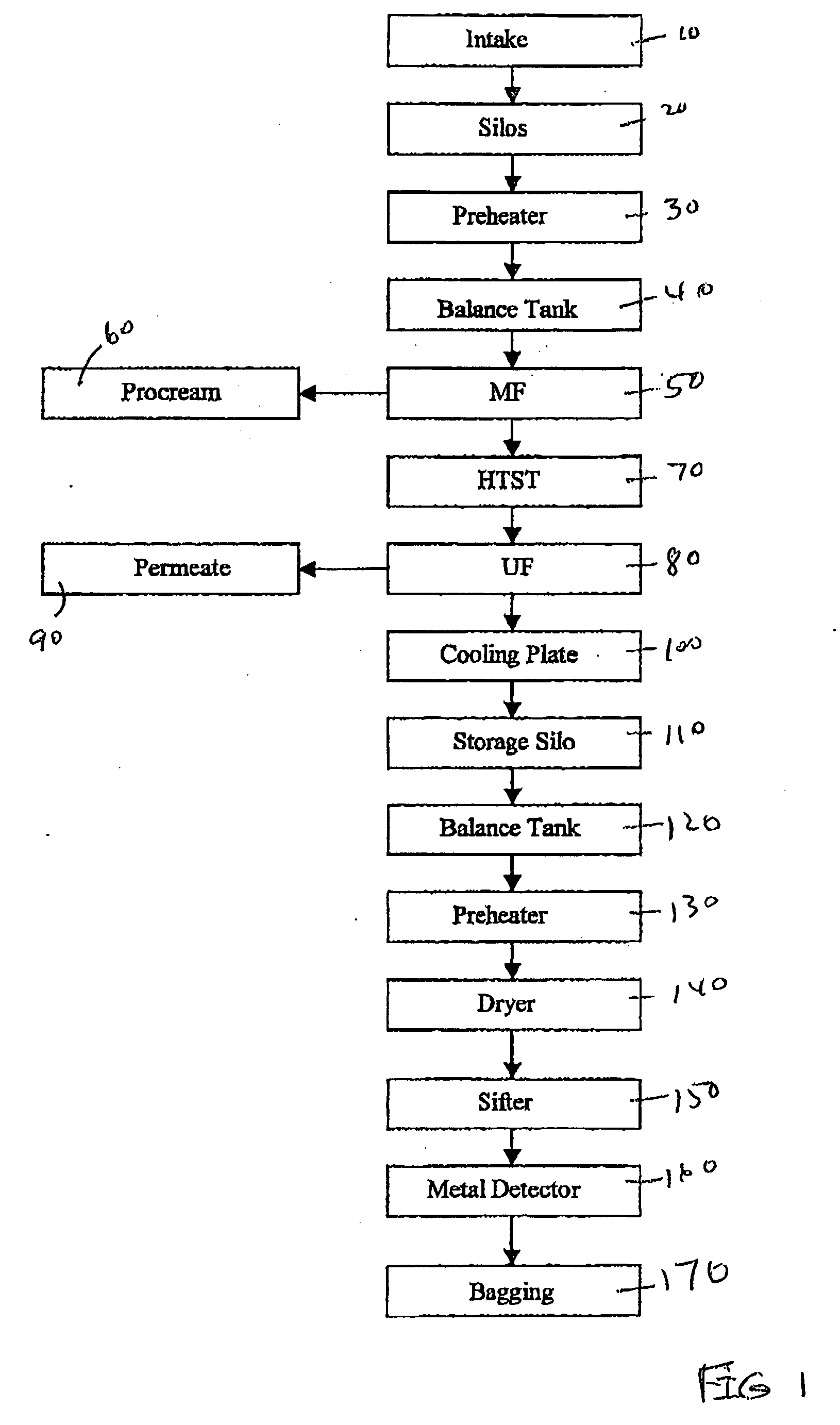
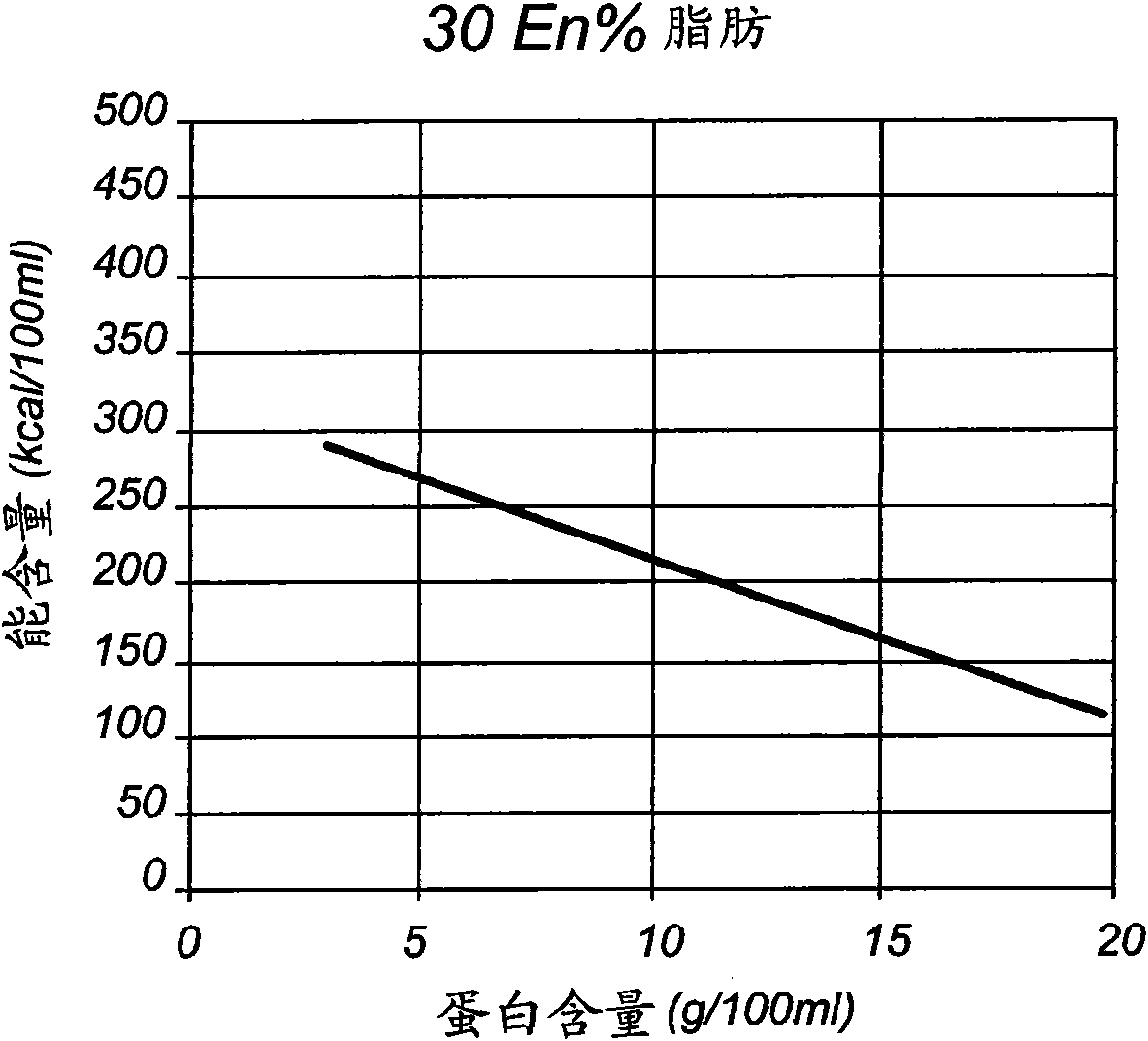
Liquid enteral nutritional composition with a low specific protein volume
InactiveCN101909468AEasy to take orallyGreat tasteMetabolism disorderOther dairy technologyDiseaseOlder people
The present invention concerns liquid enteral nutritional compositions that comprise intact protein with a low specific volume allowing the preparation of nutrition with high protein and energy contents which are particularly advantageousfor persons that are in, or recovering from, a disease state, persons that are malnourished or sportsmen and women and active elderly.
Owner:NUTRICIA
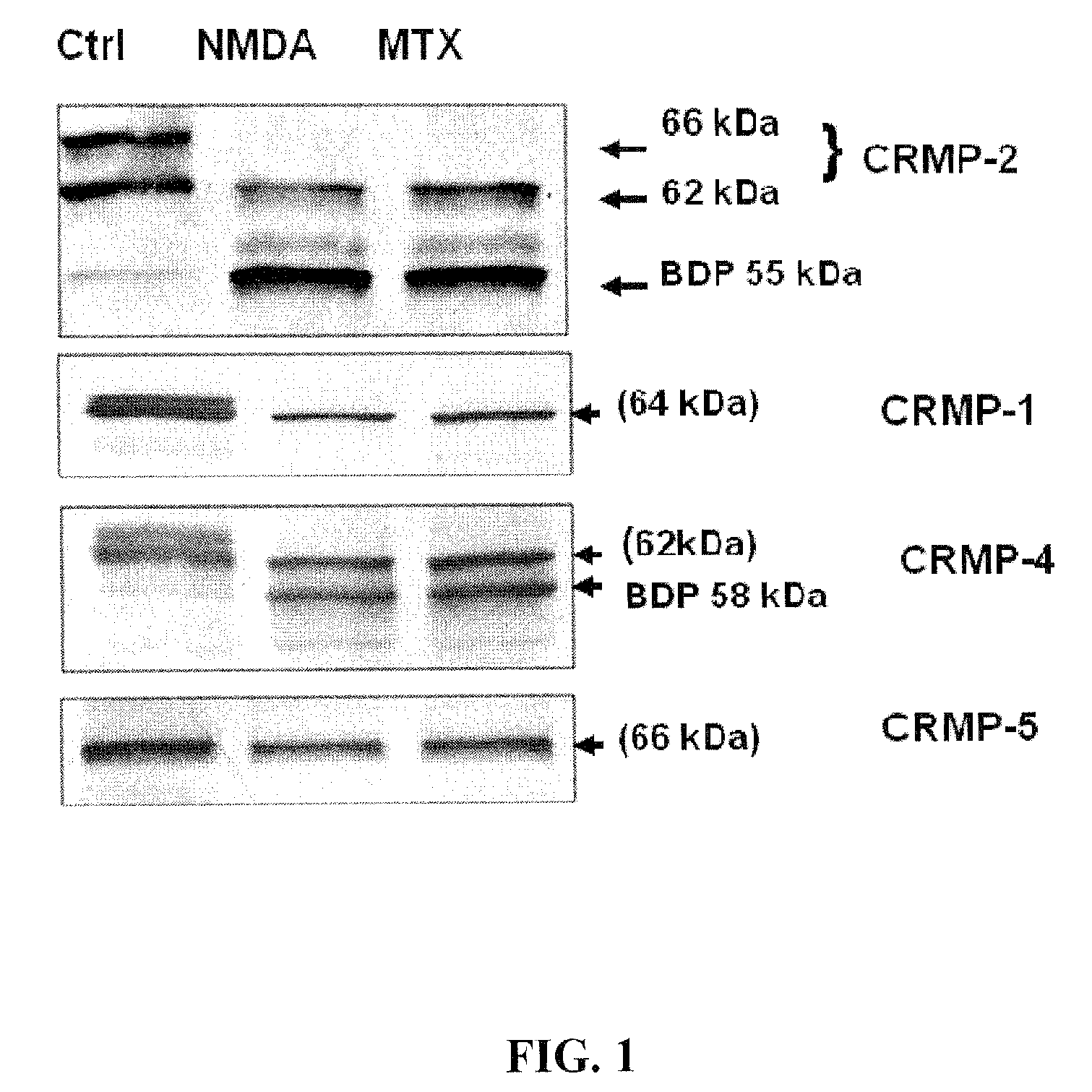
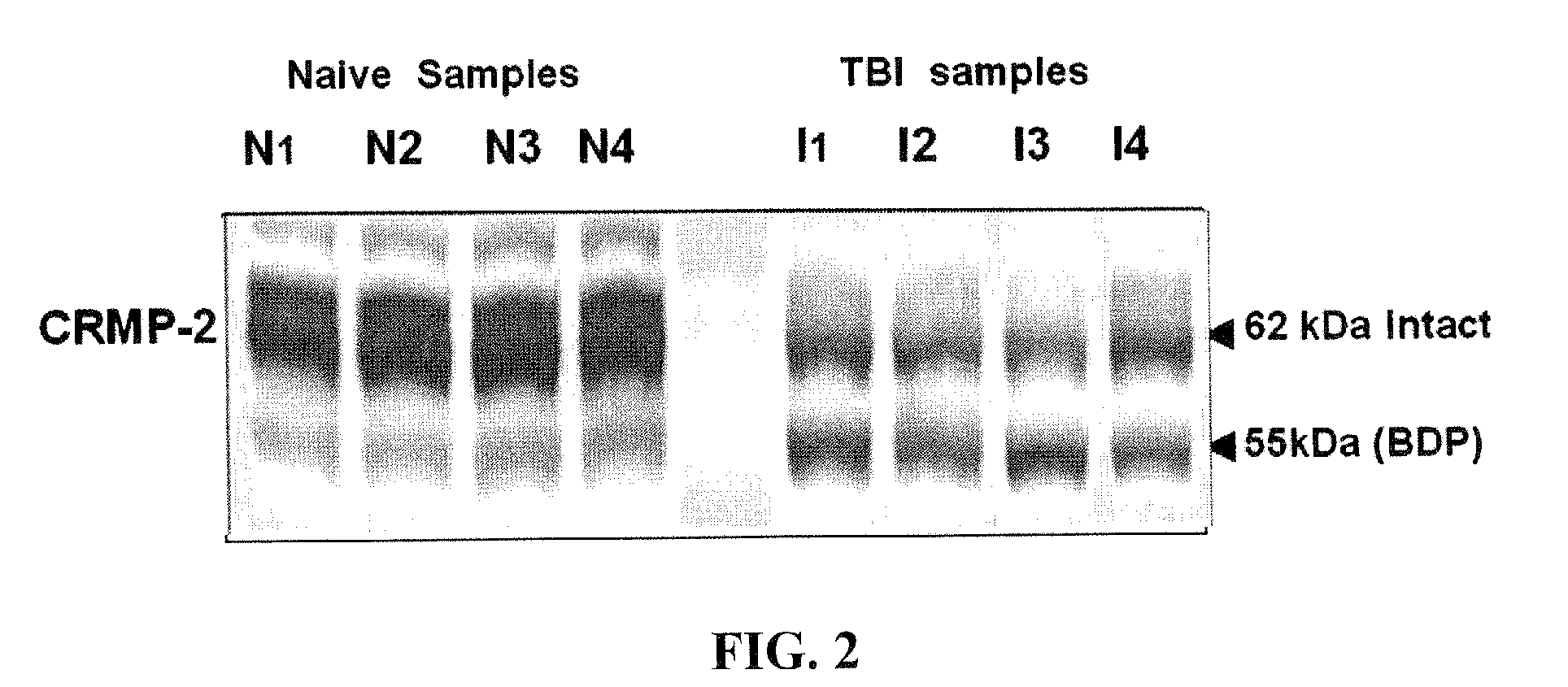
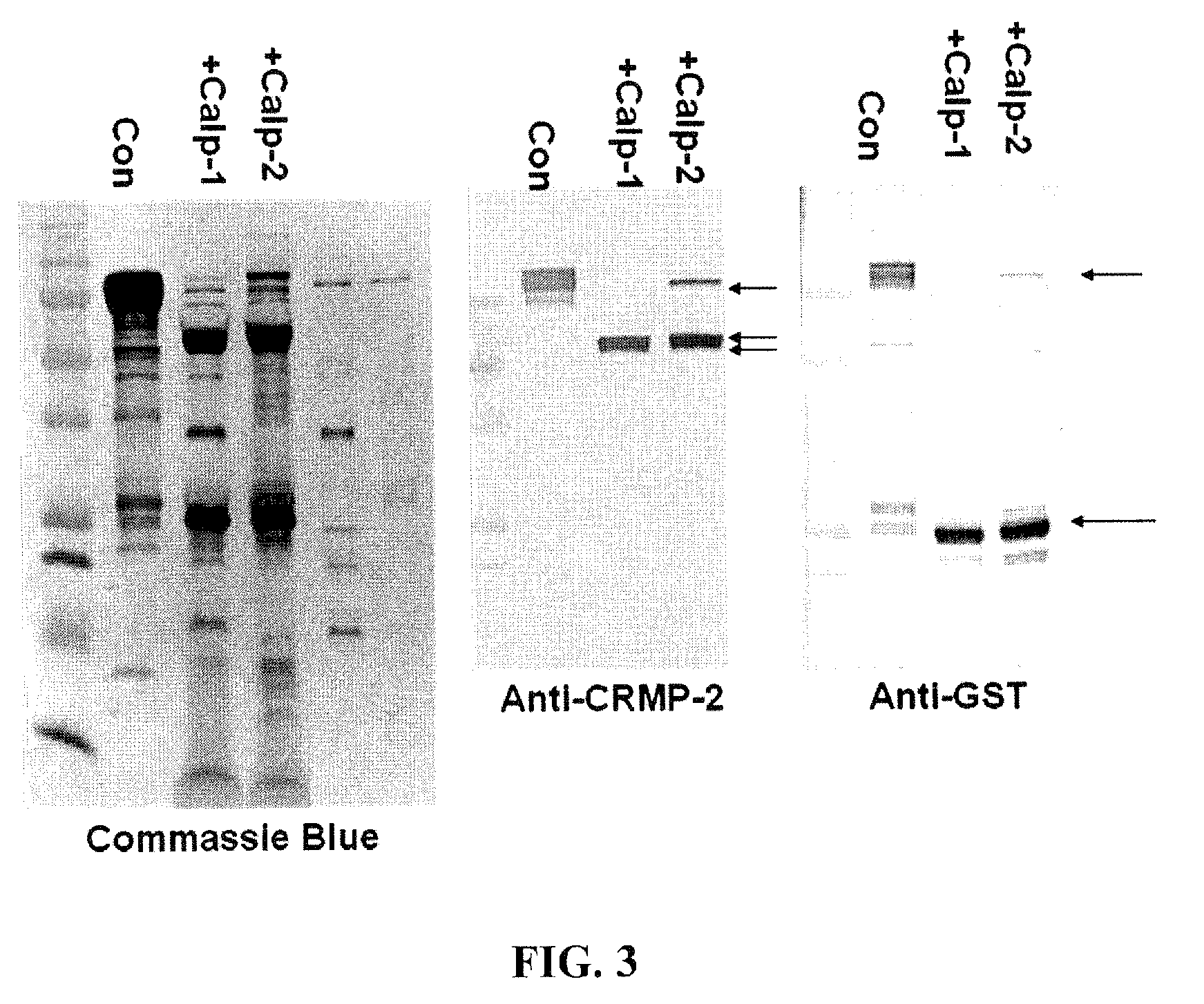
Synaptotagmin and collapsin response mediator protein as biomarkers for traumatic brain injury
ActiveUS8557526B2Information can be usedReduce in quantityDisease diagnosisBiological testingIntact proteinNeurological injury
Collapsin response mediator proteins (CRMPs) decreased in tissue and increased in biological fluids after neural injury from traumatic brain injury (TBI). Significant decreases of CRMP1, CRMP2, CRMP4 and CRMP5 were accompanied by the appearance of distinct 58 kDa (CRMP-2) or 55 kDa (CRMP-4) breakdown products from proteolytic cleavage by calpain. Synaptotagmin breakdown products were also associated with TBI and could be detected along with intact protein in human cerebral spinal fluid (CSF). Both biomarkers were detected in human biofluid and related to recovery from traumatic brain injury.
Owner:UNIV OF FLORIDA RES FOUNDATION INC +1
Nutritional composition intended for specific gastro-intestinal materation in premature mammals
Owner:SOC DES PROD NESTLE SA
Method for controlling transgene flow by using gene split
InactiveCN102220356AAvoid environmental risksVector-based foreign material introductionPlant genotype modificationIntact proteinIntein
The invention discloses a method for controlling transgene flow by using gene split, and belongs to the technical field of genetically modified organism safety. The method is characterized in that: a complete target gene is splited into an N-terminal sequence and a C-terminal sequence; the N-terminal sequence and the C-terminal sequence are respectively ligated with the gene sequence of the N-terminal splicing domain of the Intein gene and the gene sequence of the C-terminal splicing domain of the Intein gene to form a fusion gene A and a fusion gene B; the fusion gene A and the fusion gene B are transferred into a nuclear genome of a receptor plant through technical means of agrobacterium tumefaciens transformation and hybridization, or the fusion gene A is transferred into the nuclear genome while the fusion gene B is transferred into the chloroplast genome to obtain the gene-splited transgenic plant; after the two fusion genes are expressed in the transgenic plant, two inactive protein fragments are reassembled into active and complete protein in the chloroplast genome or the nuclear genome through Intein-mediated protein-splicing function. According to the method provided by the present invention, environment risk due to the transgene flow can be reduced.
Owner:THE INST OF BIOTECHNOLOGY OF THE CHINESE ACAD OF AGRI SCI
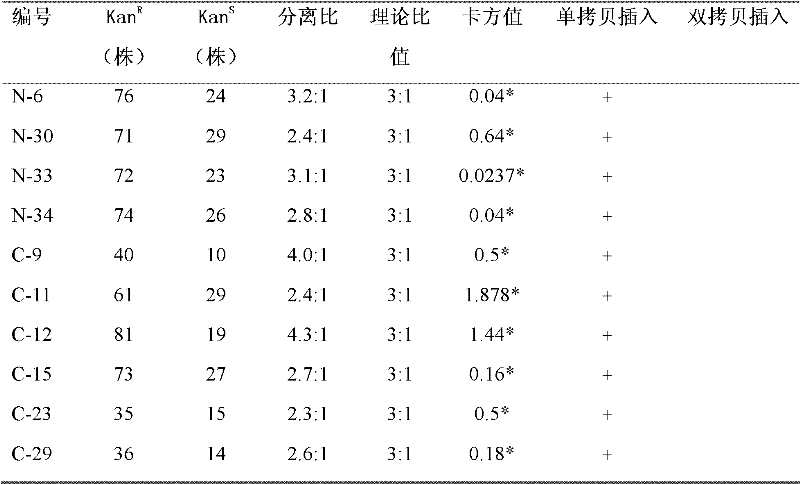
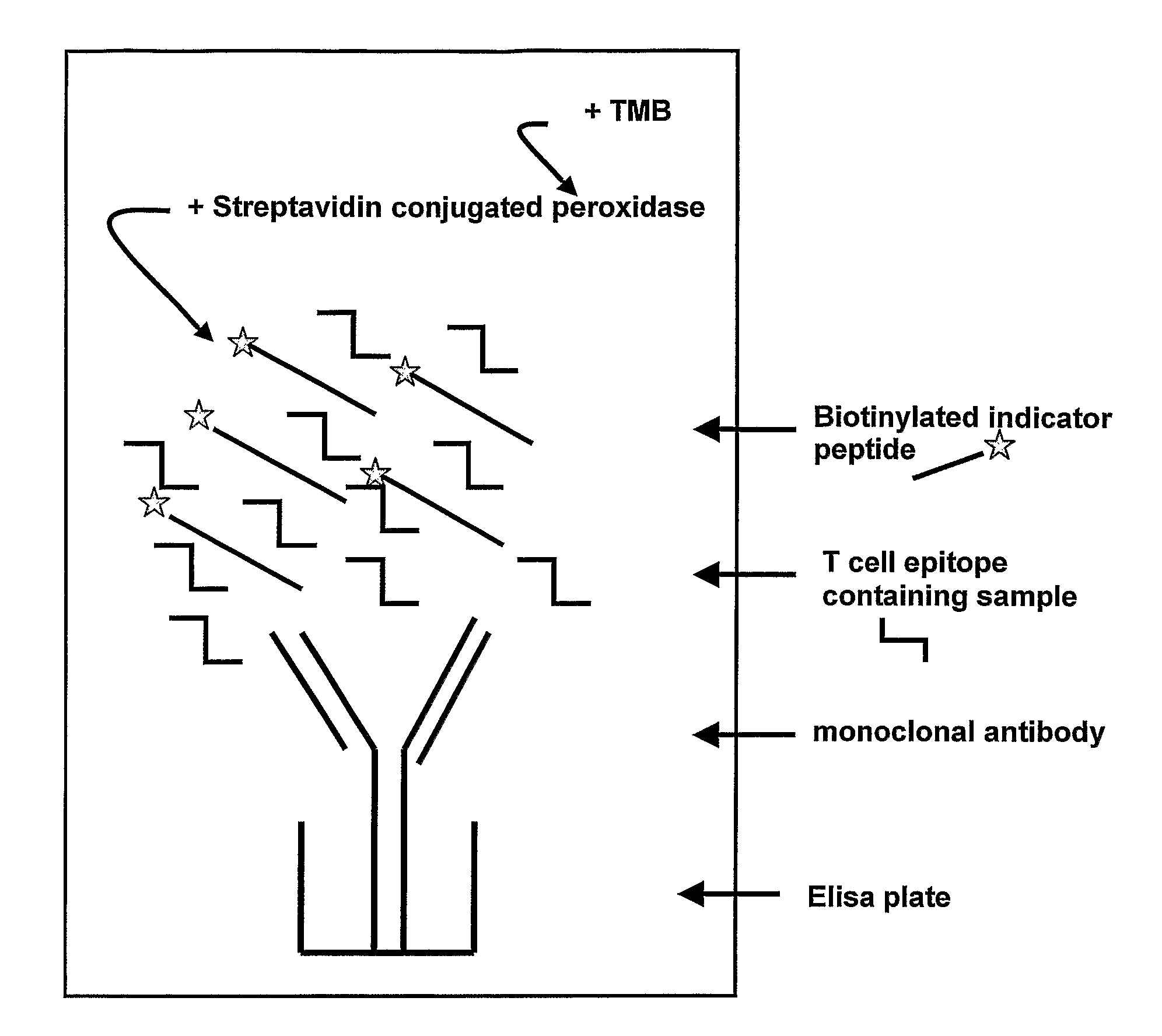
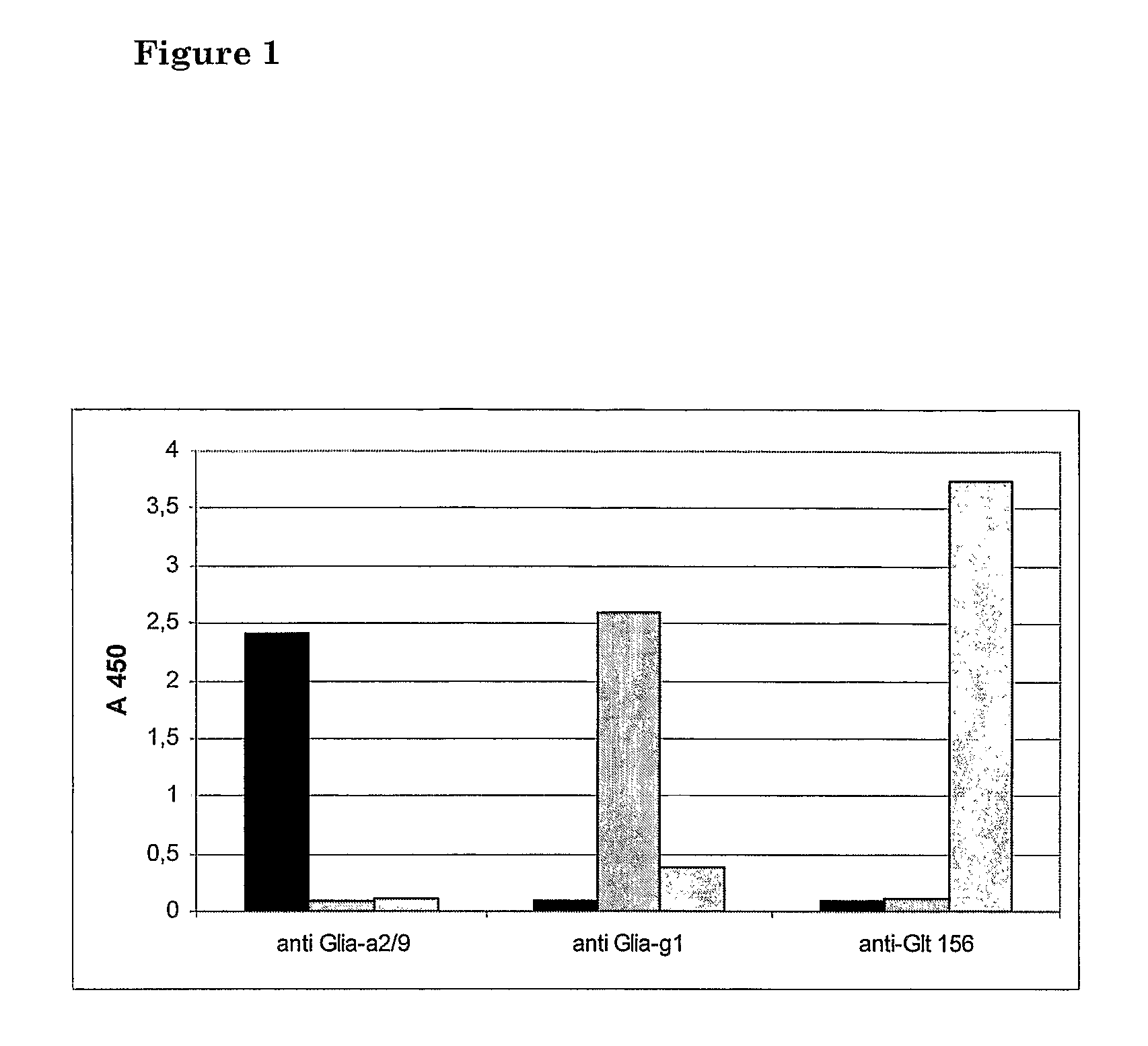
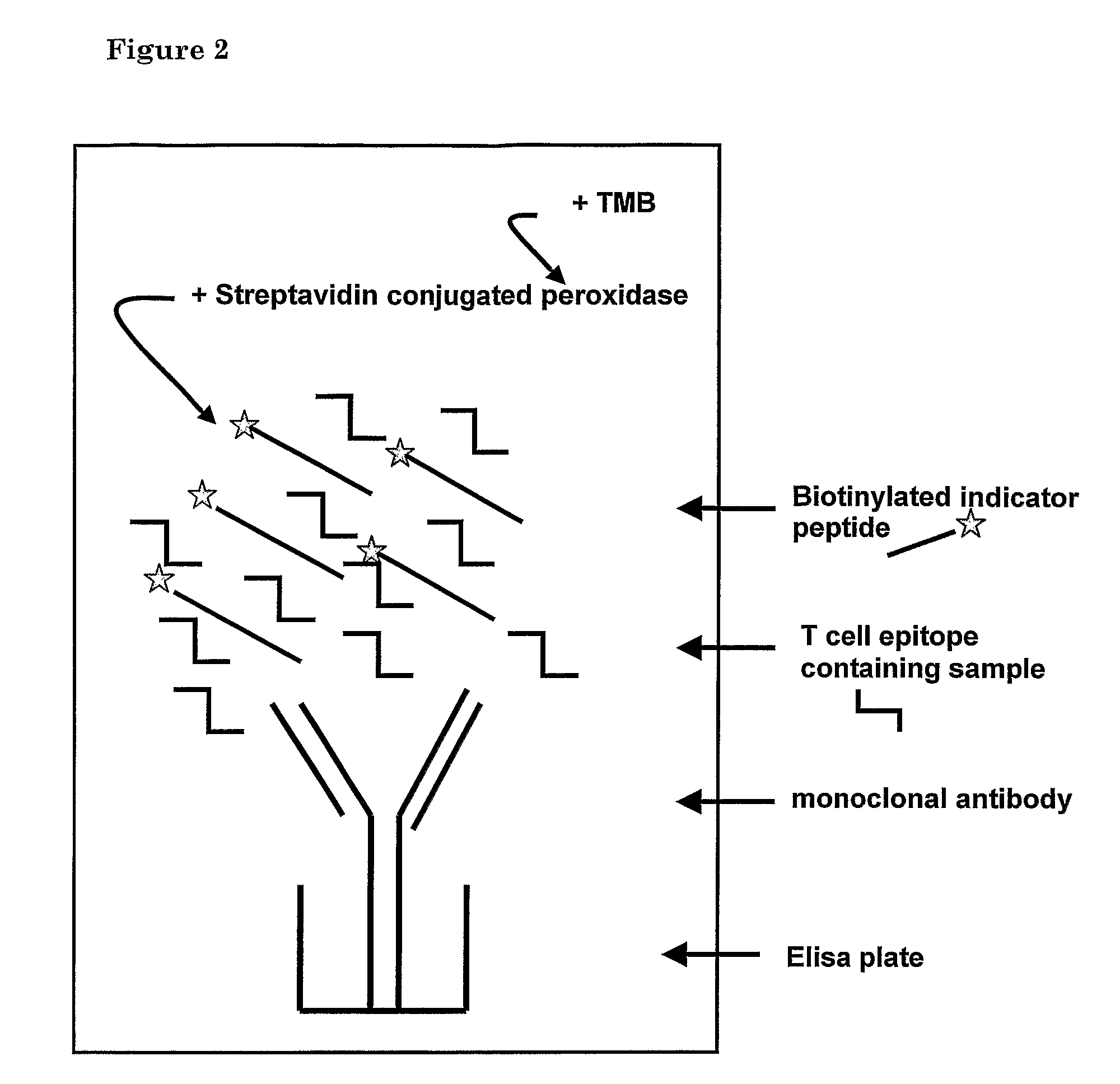
Method for detecting gluten
Provided is a mAb-based method for the detection of T cell stimulatory epitopes known to be involved in CD. The method has many advantages compared to the existing methods for the detection of gluten since it is the first method that can; (i) detect T cell stimulatory epitopes of gluten; (ii) detect the epitopes separately, (iii) detect T cell stimulatory epitopes present on gliadin and glutenin homologues present in other cereals also known to be involved in CD; and (iv) detect T cell stimulatory epitopes on both intact proteins and small protein fragments. The new method is a valuable tool in the screening of basic ingredients, semi manufactured ingredients and food products that are intended to be used in the gluten free diet of CD patients. Moreover the new method can also be used for the screening of cereals and different wheat varieties for the level of toxicity for CD patients. Thereby the method can help in the selection of cereals and wheat varieties with low toxicity which might form the basis for future breeding programs. In the future these cereals will be used for the production of safe food for CD patients.
Owner:ACADEMISCH ZIEKENHUIS BIJ DE UNIV VAN AMSTERDAM ACADEMISCH MEDISCH CENT
Nutrition bar
Owner:UNILEVER NV
Proteins that stimulate the secretion of satiety hormones
InactiveUS20110217380A1Improve the level ofFacilitated releasePowder deliveryPeptide/protein ingredientsIntact proteinDelivery vehicle
The invention is in the field of weight management, in particular in the field of weight management by influencing the mechanisms of body-weight regulation. Intact pea protein and intact wheat protein were found to be effective in reducing appetite or inducing or increasing satiety when brought into contact with their receptors in the duodenum. Since it is known that intact proteins hydrolyse in the gastrointestinal tract, intact pea protein and intact wheat protein will not exhibit their satiating effect when ingested in a conventional oral preparation. Therefore, special care should be taken to deliver the intact proteins to the duodenum in order for them to arrive there intact. One object of the invention may therefore be achieved by incorporating the intact protein in an enteric delivery vehicle.
Owner:MAASTRICHT UNIVERSITY
Storage method of sweet potatoes
InactiveCN107114467APrevent rotInhibition of respirationFruits/vegetable preservation by irradiation/electric treatmentFruits/vegetable preservation by dehydrationAgricultural scienceIntact protein
The present invention mainly relates to the technical field of planting and discloses a storage method of sweet potatoes. The method comprises drying, bacterium preventing, bud inhibiting and storing. The method is simple and can be subjected to batch operations. After being stored for 10 months, an intact rate of the sweet potatoes reaches 97.4 %, a germination rate is only 0.46%, and economic income is increased by 12.3%. After the sweet potatoes are harvested, the harvested sweet potatoes are spray-washed using a washing solution, surface bumps of the sweet potatoes are reduced, surface soil is removed, and the washing solution comprises a variety of plant extracts, inhibits growth of infectious microbes, promotes wound healing and avoids sweet potato rots; and after washing, the sweet potatoes are placed in a warehouse to be subjected to a drying, and after a small amount of water losing, respiration functions and overall metabolisms of the sweet potatoes are inhibited, nutrients of the sweet potatoes are retained, germination of the sweet potatoes is inhibited, and a storage time of the sweet potatoes is extended.
Owner:蚌埠市乔峰农业蔬菜专业合作社
Mass spectrometric methods and kits to identify a microorganism
ActiveUS20190212338A1Rapidly and efficiently removedIncrease signal strengthBiological testingMicroorganismComplete protein
The present invention includes a novel method and system for identification of microorganisms in samples that proteins and other biological material from non-microorganism sources (e.g., proteins of mammalian origin) that can interfere with identification of the microorganisms. The methods and systems described herein include use of a single-use chromatography medium to purify intact proteins prior to mass spectrometry analysis. The chromatography medium and the methods described herein can rapidly and efficiently remove of a substantial portion of interfering biological material (e.g., mammalian proteins) from a crude cell lysate while preserving high signal strength and removing enough of the interfering protein(s) to allow for identification of the microorganism(s) by mass spectrometry analysis.
Owner:THERMO FISCHER SCI OY
Nutritional Compositions Containing a Peptide Component and Uses Thereof
The present disclosure relates to nutritional compositions including a protein equivalent source including a peptide component comprising selected peptides from Tables 1 and 2 disclosed herein. The protein equivalent source may further include intact protein, hydrolyzed protein, including partially hydrolyzed protein, or combinations thereof. The disclosure further relates to methods of reducing the incidence of autoimmune disease and / or diabetes mellitus by providing said nutritional compositions to a target subject, which includes a pediatric subject.
Owner:MEAD JOHNSON NUTRITION
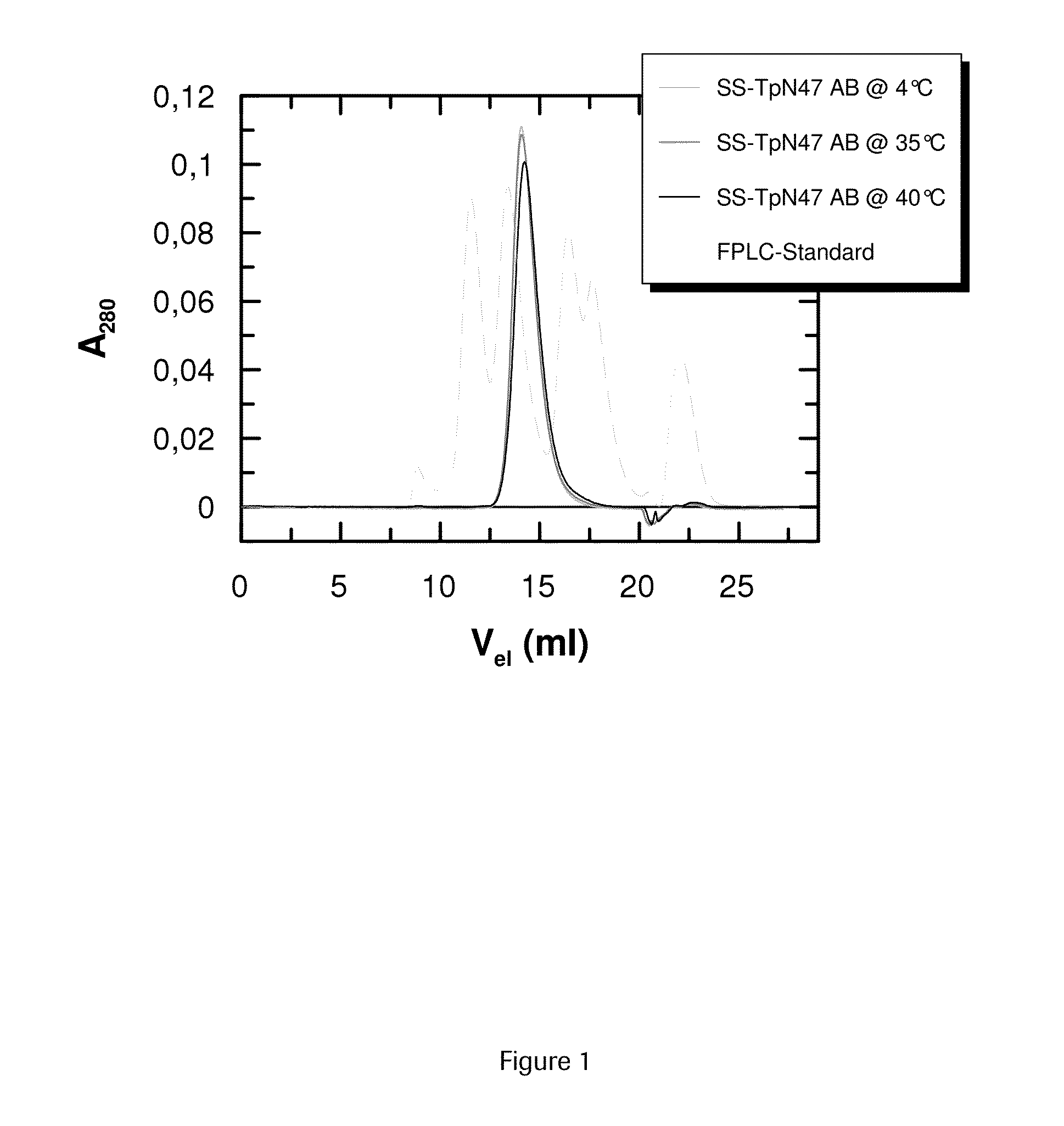
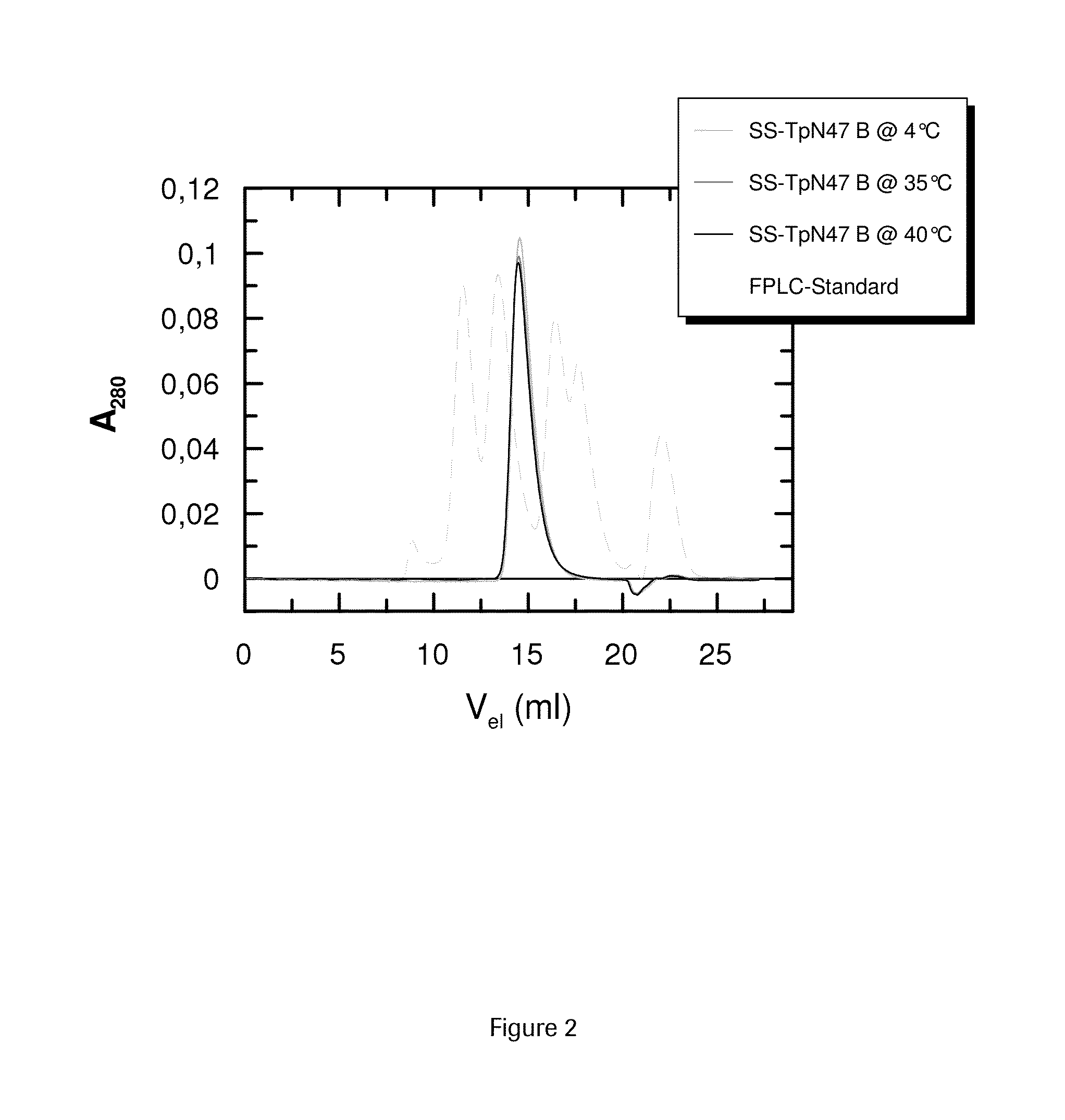
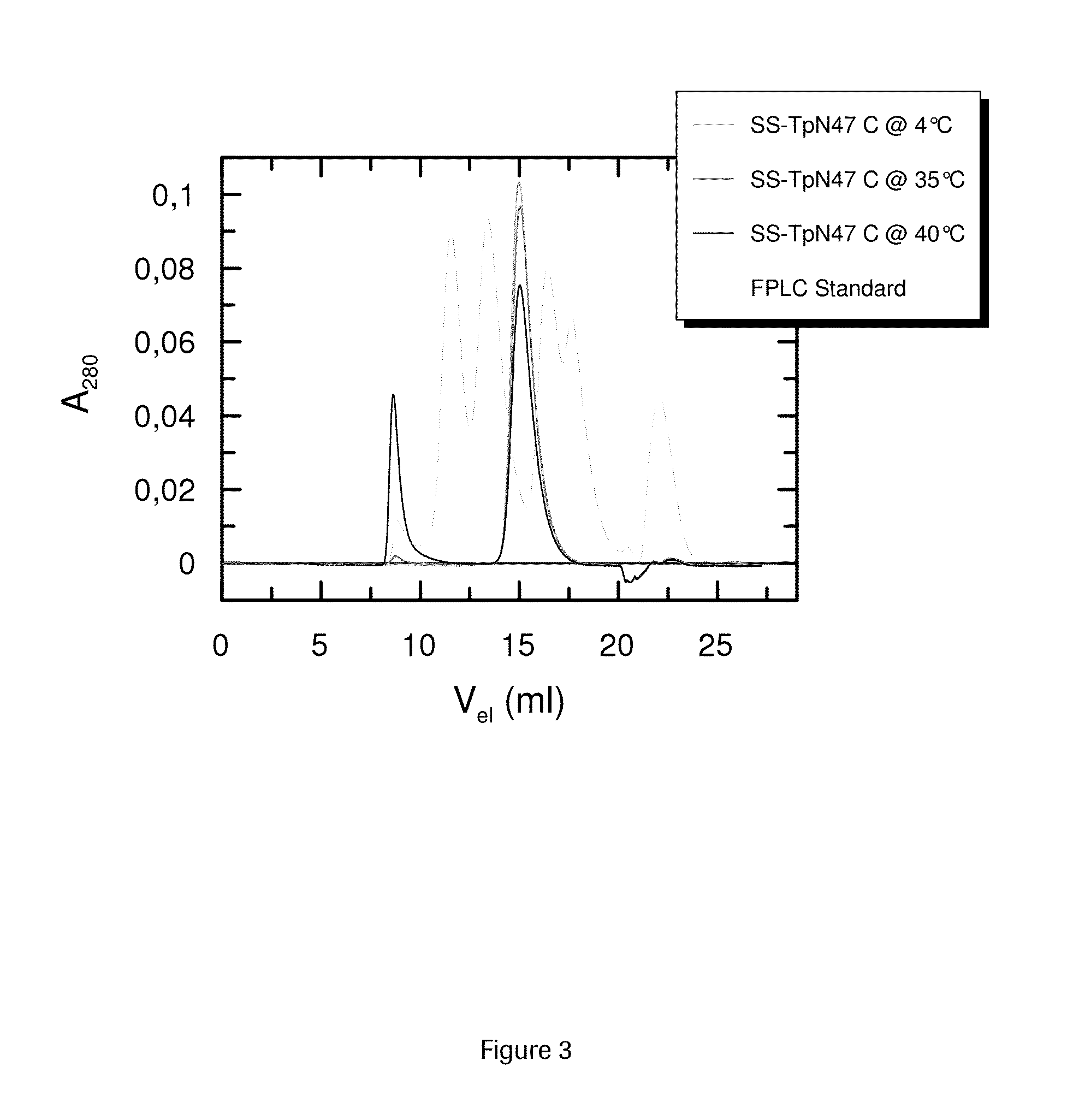
SOLUBLE IMMUNOREACTIVE TREPONEMA PALLIDUM TpN47 ANTIGENS
ActiveUS20150079604A1Improve solubilityAntibody mimetics/scaffoldsPeptide preparation methodsAntigenSpiroplasma
The invention concerns soluble variants of Treponema pallidum antigen 47 (TpN47 antigen) comprising at least domain B, or at least domains A and B, optionally domain D of the complete TpN47 protein molecule with the proviso that all antigens lack domain C (amino acid residues 224 to 351) of TpN47. The Tpn47 antigens can be fused to a chaperone. Moreover, the invention covers DNA encoding the antigens, a method of producing these antigens as well as the use of these antigens in an immunodiagnostic assay for the detection of antibodies against Treponema pallidum in an isolated sample.
Owner:ROCHE DIAGNOSTICS OPERATIONS INC
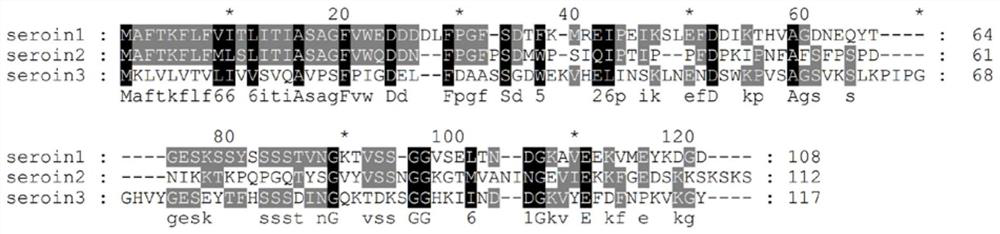
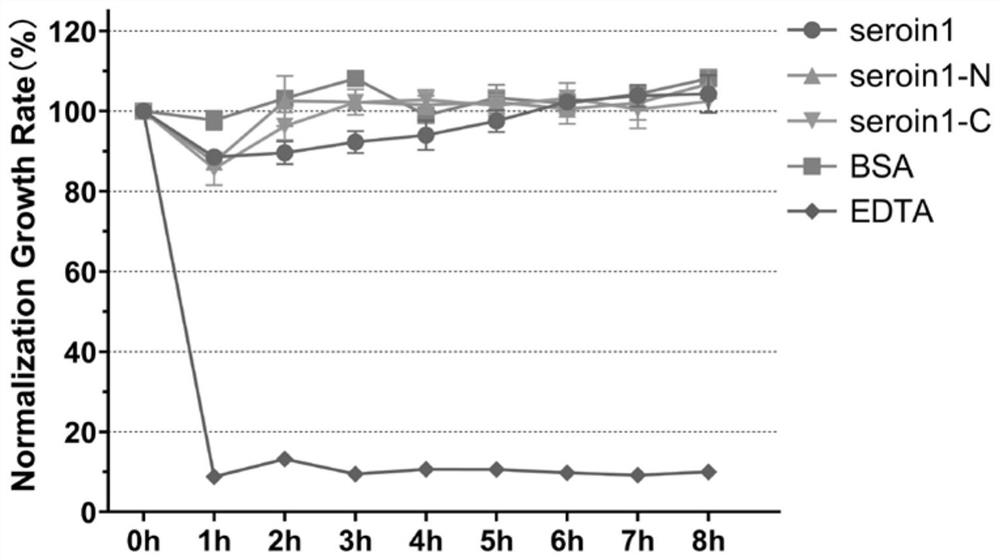
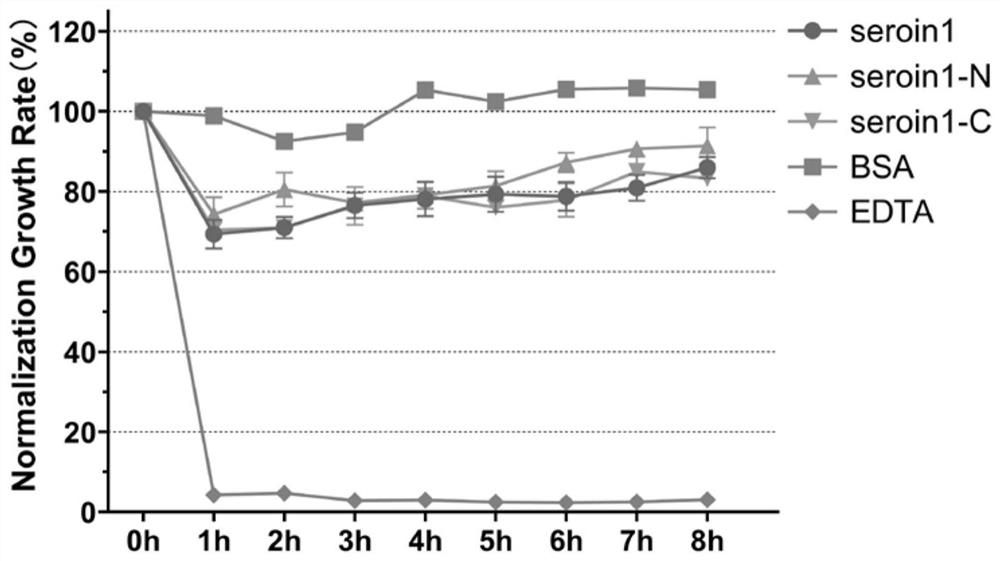
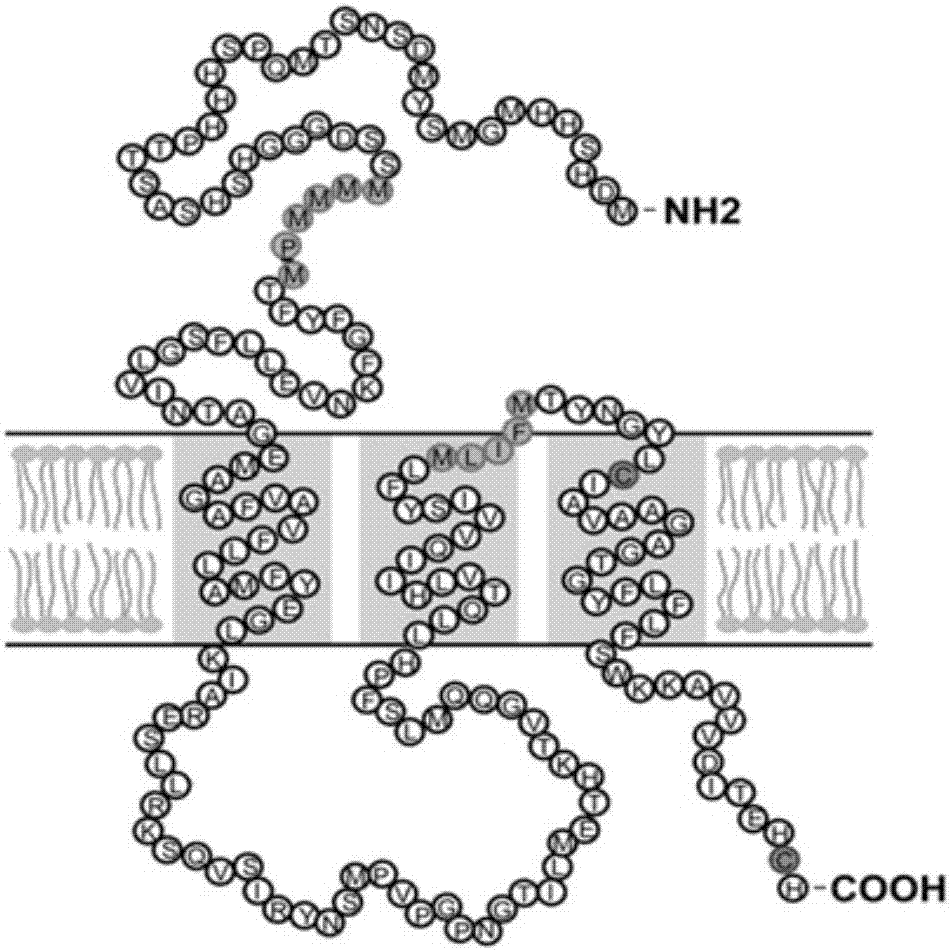
Truncated body of Seroin protein and application of truncated body
ActiveCN112375131AHas antibacterial activityHigh antibacterial activityAntibacterial agentsPeptide/protein ingredientsComplete proteinIntact protein
The invention discloses a truncated body of a Seroin protein and application of the truncated body. The truncated body of the Seroin protein is an N-end or C-end fragment of Seroin 1, Seroin 2 or Seroin 3. The truncated polypeptide has antibacterial activity and can be used for resisting Gram-positive bacteria and Gram-negative bacteria, the molecular weight of protein is greatly reduced through truncation, and therefore the cost and difficulty of synthesizing the polypeptide by a chemical method are reduced, and compared with a previous complete protein production technology, the truncated body of a Seroin protein has the obvious advantages of low production cost, high activity, high stability, simplicity, convenience, quickness and the like.
Owner:SOUTHWEST UNIV
Method for screening multi-transmembrane region protein antibody and application of multi-transmembrane region protein antibody
InactiveCN107982526AIncrease concentrationEfficient identificationCell receptors/surface-antigens/surface-determinantsSerum immunoglobulinsAntigenComplete protein
The invention provides a method for screening a multi-transmembrane region protein antibody and an application of the multi-transmembrane region protein antibody. The method comprises the following steps: by taking a multi-transmembrane region protein as an antigen, screening an antibody for complete proteins, and by taking a baculovirus with the multi-transmembrane region protein as an antigen, identifying the antibody for an extracellular region of the multi-transmembrane region protein. The invention further relates to an antibody prepared by using the method and an application of the antibody.
Owner:深圳市中瑞牛津生命科学技术有限公司
Fusion protein molecular weight analysis method
ActiveCN105628927AAccurate determination of molecular weightBiological testingIntact proteinChemistry
The present invention discloses a fusion protein molecular weight analysis method, and specifically relates to a method for carrying out complex glycosylated fusion protein accurate molecular weight analysis by using ESI-Q-TOF, particularly to a sample pre-treatment method for removing fusion protein N-glycan and sialic acid by using specific glycan removing enzyme, wherein the treated fusion protein is separated through a salt removing column, the effluent is broken through an ESI source to obtain continuous multi-charged ions, and tandem Q-TOF mass spectrometry analysis and deconvolution calculation are performed to obtain the complete protein accurate molecular weight of the O-glycan core isomer. With the method of the present invention, the high inhibition of the N-glycan and / or sialic acid on the fusion protein ionization efficiency can be overcome, and the complete protein molecular weight of the O-glycan fusion protein can be accurately determined.
Owner:SUNSHINE GUOJIAN PHARMA (SHANGHAI) CO LTD
Nutritional compositions containing a peptide component and uses thereof
The present disclosure relates to nutritional compositions including a protein equivalent source including a peptide component comprising selected peptides from Tables 1 and 2 disclosed herein. The protein equivalent source may further include intact protein, hydrolyzed protein, including partially hydrolyzed protein, or combinations thereof. The disclosure further relates to methods of reducing the incidence of autoimmune disease and / or diabetes mellitus by providing said nutritional compositions to a target subject, which includes a pediatric subject.
Owner:MEAD JOHNSON NUTRITION
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com