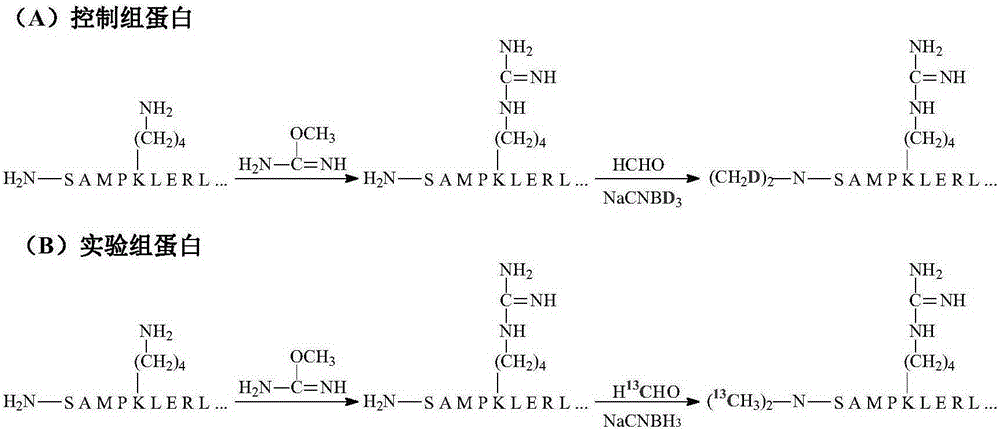Quantitative analysis method for intact protein under different physiological or pathological conditions
A quantitative analysis and protein technology, applied in the field of proteomics and systems biology, can solve the problems of limited application range, and achieve the effect of high labeling efficiency and high accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
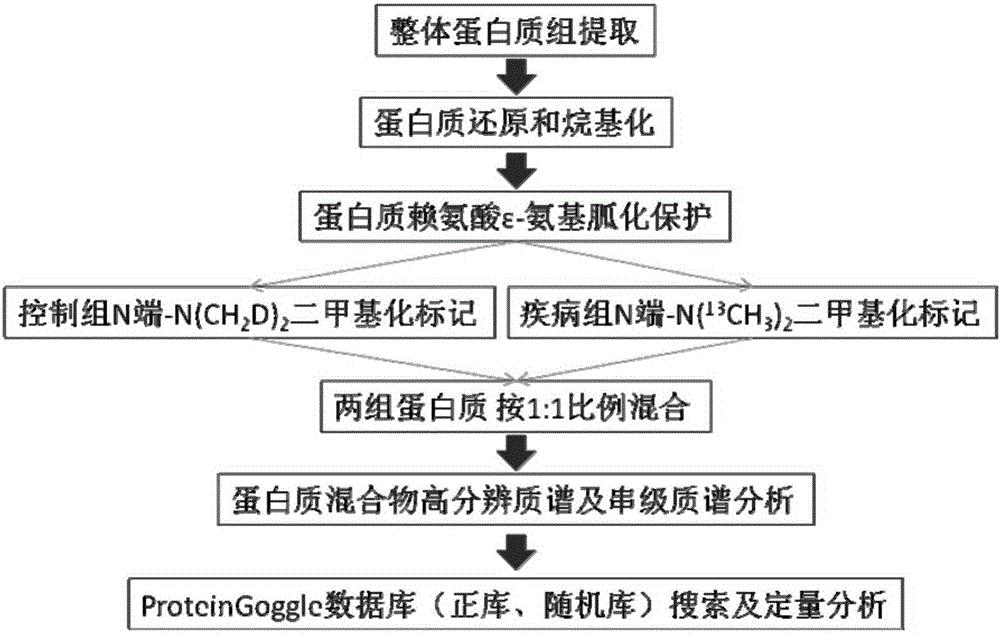
[0021] A method for the quantitative analysis of whole proteins under different physiological or pathological conditions, using such as figure 1 The overall process shown includes the following steps:
[0022] (1) The overall protein of two different physiological or pathological conditions (take the control group and the disease group as an example) is first reduced with dithiothreitol and alkylated with iodoacetamide;
[0023] (2) The above two groups of alkylated proteins were uniformly guanidinated with O-methylisourea (2M, 100mM sodium bicarbonate solution) on the ε-amino group on the lysine residue in the protein sequence (use 2M NaOH to adjust the pH to 11, 65°C), quenched with 10% (v / v) trifluoroacetic acid solution after 15 minutes of reaction;
[0024] (3) The two groups of proteins after protection were respectively treated with formaldehyde and sodium deuterated cyanoborohydride, 13 C-labeled formaldehyde and sodium cyanoborohydride are used to label the N-termin...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com