Prediction algorithm for recognizing tyrosine posttranslational modification sites
A post-translation modification and prediction algorithm technology, applied in computing, special data processing applications, instruments, etc., can solve problems such as few training samples, unsatisfactory prediction performance, and long time consumption, and achieve the effect of improving prediction ability.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
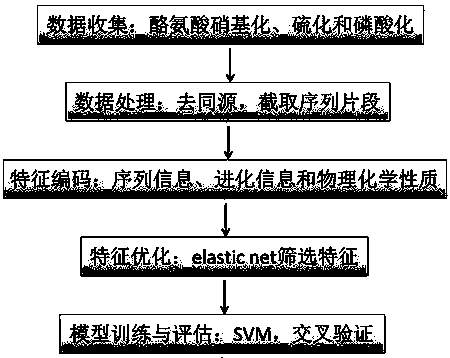
[0061] a kind of like figure 1 The specific steps of the prediction algorithm for identifying tyrosine post-translational modification sites shown are as follows:
[0062] 1) Data Collection
[0063] Collect data on tyrosine nitration, sulfuration and phosphorylation post-translational modifications from protein databases and related literature in recent years. The protein database is at least one of the PhosphoSitPlus database, UniProtKB / Swiss-Prot database, SYSPTM database and dbPTM database species, the PhosphoSitPlus database only collects data that can undergo tyrosine phosphorylation with relevant kinase annotations;
[0064] 2) Data processing
[0065] Use the CD-HIT program to remove the homology collected from several different protein databases, that is, highly homologous protein sequences with the same or similarity greater than 30%, to obtain non-redundant tyrosine nitrosylation, sulfuration and phosphorylation modifications The positive sample data set and the ...
Embodiment 2
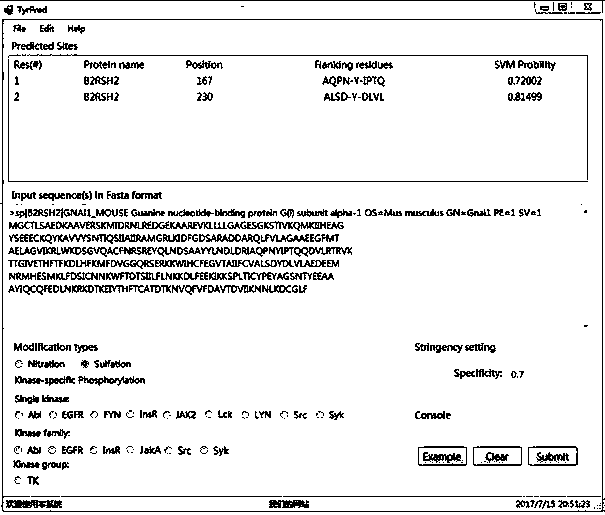
[0092] The prediction software platform TyrPred was applied to predict the tyrosine nitration site and tyrosine sulfuration site of the protein named "B2RSH2".
[0093] The prediction software is TyrPred, a prediction software platform developed by using MATLAB software and C# programming language to build the optimal model based on SVM. The prediction software platform TyrPred, after the user submits at least one unknown protein sequence in FASTA format and selects the type of post-translational modification to be predicted, will efficiently return the prediction information of potential tyrosine post-translational modification sites, and realize simultaneous analysis of the complete protein. High-throughput prediction of tyrosine nitration, sulfuration and phosphorylation sites, prediction information includes protein name, modification site position, flanking residues of modification site and SVM probability value.
[0094] To predict the nitration site of the protein seque...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com