Peronia verruculata microsatellite marking and screening method
A technology of microsatellite markers and screening methods, which is applied in the field of microsatellite markers and screening of purple warts, can solve the problems of genetic diversity of unseen molecular markers, and achieve the effect of low cost
Inactive Publication Date: 2016-08-17
SHANGHAI OCEAN UNIV
View PDF1 Cites 4 Cited by
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
[0004] As a transitional species of marine invertebrates extending to land, the clamshell shellfish of the family Sulphuridae is considered to be the best representative for the study of marine shellfish radiating to land. In the report of genetic diversity, the 7 microsatellite marker sites developed by high-throughput sequencing technology in the present invention not only provide basic data for the study of the genetic diversity of purple verrucoids, but also can be used for the study of the genetic diversity of sulphuridae. In the study of phylogenetic relationships
Method used
the structure of the environmentally friendly knitted fabric provided by the present invention; figure 2 Flow chart of the yarn wrapping machine for environmentally friendly knitted fabrics and storage devices; image 3 Is the parameter map of the yarn covering machine
View moreImage
Smart Image Click on the blue labels to locate them in the text.
Smart ImageViewing Examples
Examples
Experimental program
Comparison scheme
Effect test
Embodiment
[0038] 1.1 Experimental materials
the structure of the environmentally friendly knitted fabric provided by the present invention; figure 2 Flow chart of the yarn wrapping machine for environmentally friendly knitted fabrics and storage devices; image 3 Is the parameter map of the yarn covering machine
Login to View More PUM
 Login to View More
Login to View More Abstract
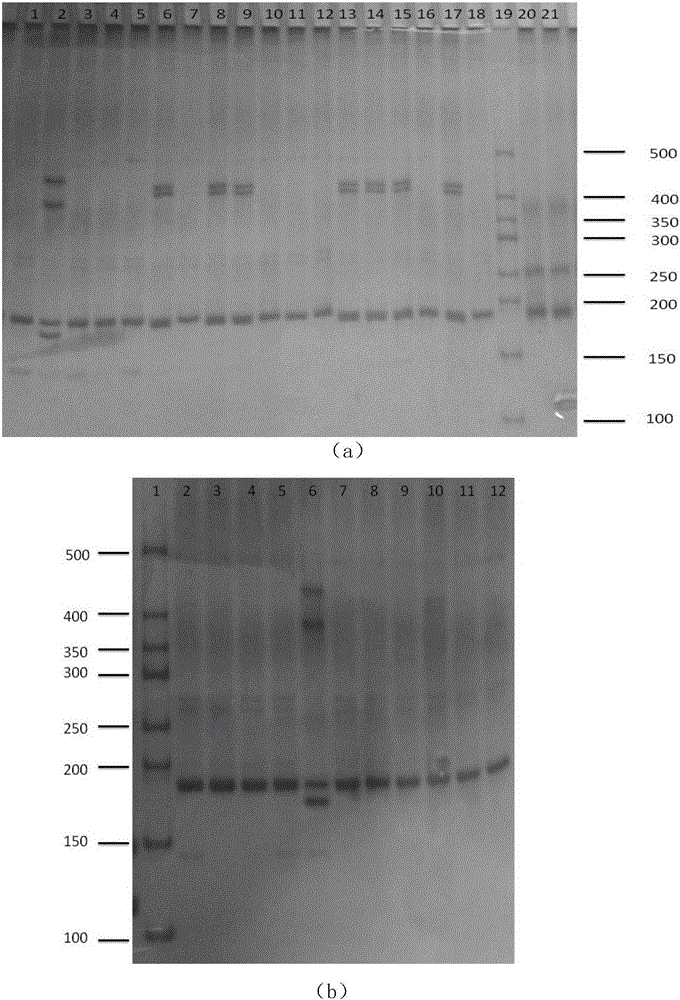
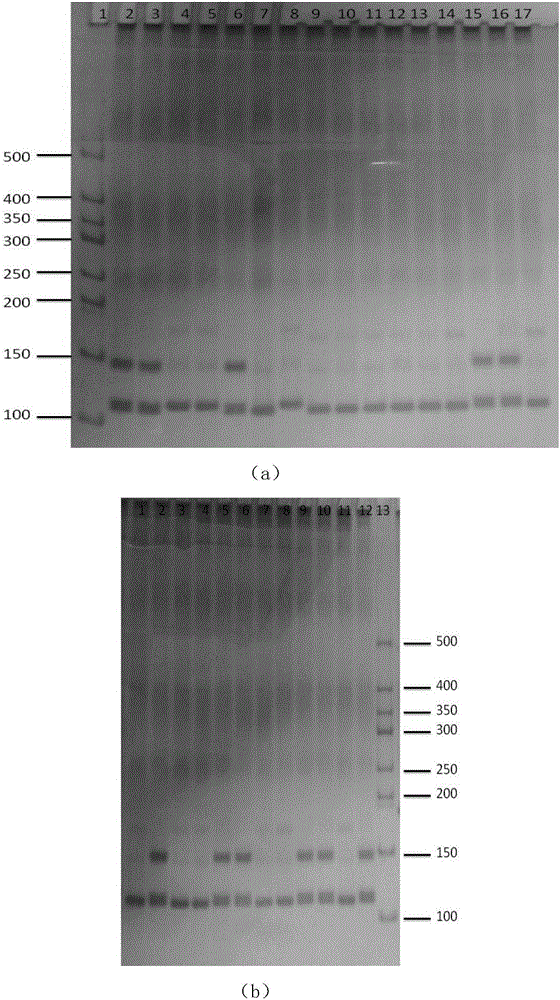
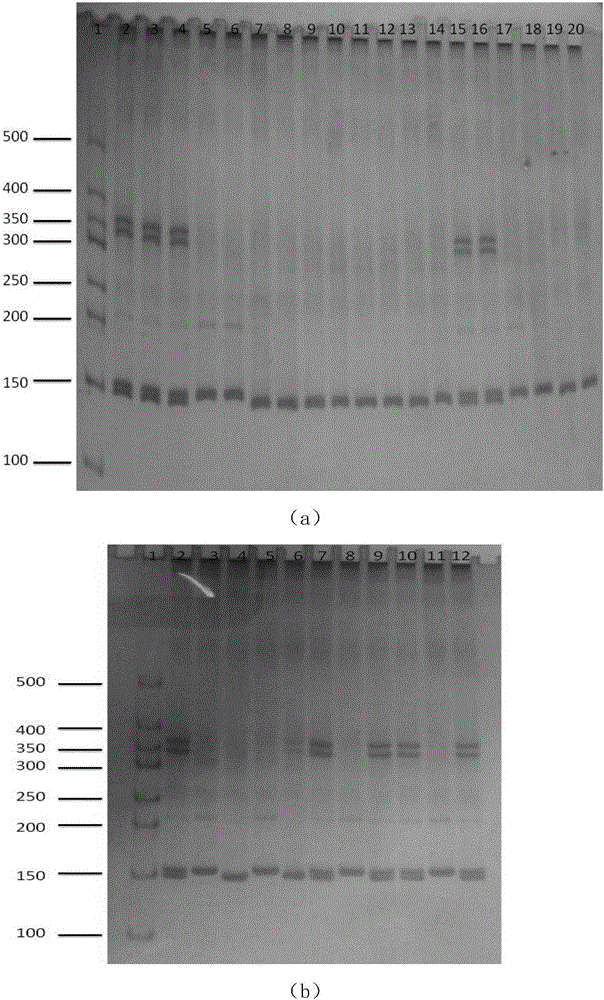
The invention discloses a peronia verruculata microsatellite marking and screening method. The method includes the steps that a peronia verruculata transcriptome sequencing library is built for Illumina HiseqTM2000 paired-end sequencing; based on a sequence obtained through sequencing, analysis and screening are conducted on microsatellite sites through MISA; screening is conducted again through SSRHunter1.3, and it is guaranteed that a front lateral wing and a rear lateral wing of the sequence have enough length for primer design; by means of an SSR primer, genome DNA of a peronia verruculata population is subjected to PCR amplification; amplification products are detected through polyacrylamide gel electrophoresis, and genotypes of the sites are determined according to different migration distances of the amplification products during population detection. Seven polymorphic microsatellite marks are obtained so as to obtain a polymorphic map of peronia verruculata genetic variation. The method is simple and quick, results are true and reliable, and the developed SSR marks can be used for phylogenetic study between peronia verruculata population genetics and onchidium shellfishes.
Description
technical field [0001] The invention belongs to the field of microsatellite molecular markers, in particular to a purple verrucous sulfonic acid microsatellite marker and a screening method. Background technique [0002] The genomes of organisms, especially the genomes of higher organisms, contain a large number of repetitive sequences, which can be divided into tandem repetitive sequences and interspersed repetitive sequences according to the distribution of repetitive sequences in the genome. Among them, the tandem repeat sequence is formed by connecting the first positions of related repeat units and arranging them in series. There are two main types of tandem repeats found so far: one is composed of functional genes (such as rRNA and histone genes); the other is composed of non-functional sequences. Tandem repeats can be classified into satellite DNA, microsatellite DNA, minisatellite DNA and the like according to the length of the repeating unit of the repeating sequen...
Claims
the structure of the environmentally friendly knitted fabric provided by the present invention; figure 2 Flow chart of the yarn wrapping machine for environmentally friendly knitted fabrics and storage devices; image 3 Is the parameter map of the yarn covering machine
Login to View More Application Information
Patent Timeline
 Login to View More
Login to View More IPC IPC(8): C12Q1/68C12N15/11
CPCC12Q1/6869C12Q1/6888C12Q2600/156C12Q2535/122C12Q2525/151
Inventor 史艳梅顾冰宁许国绿吴欣沈和定
Owner SHANGHAI OCEAN UNIV
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com