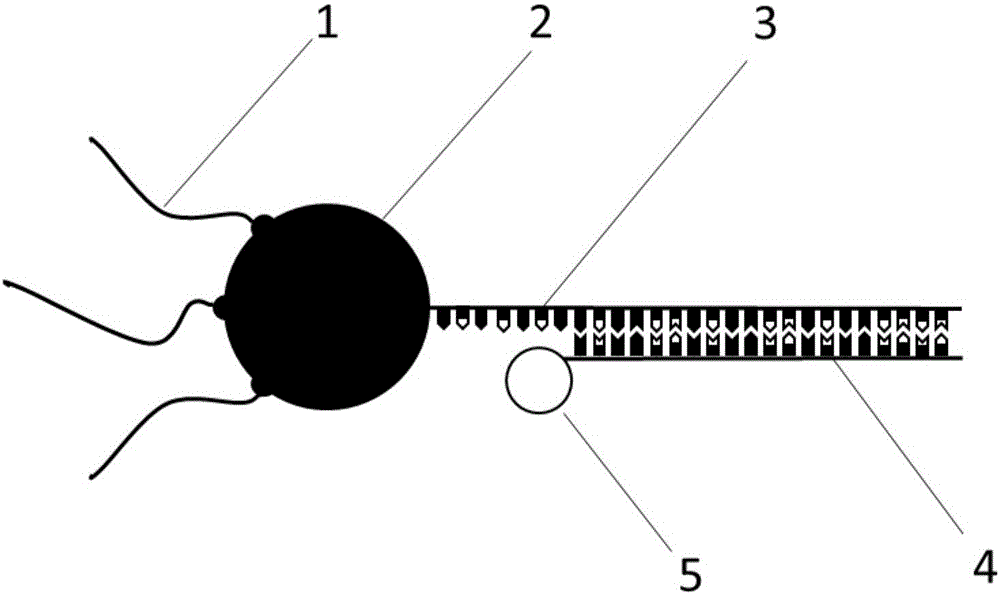
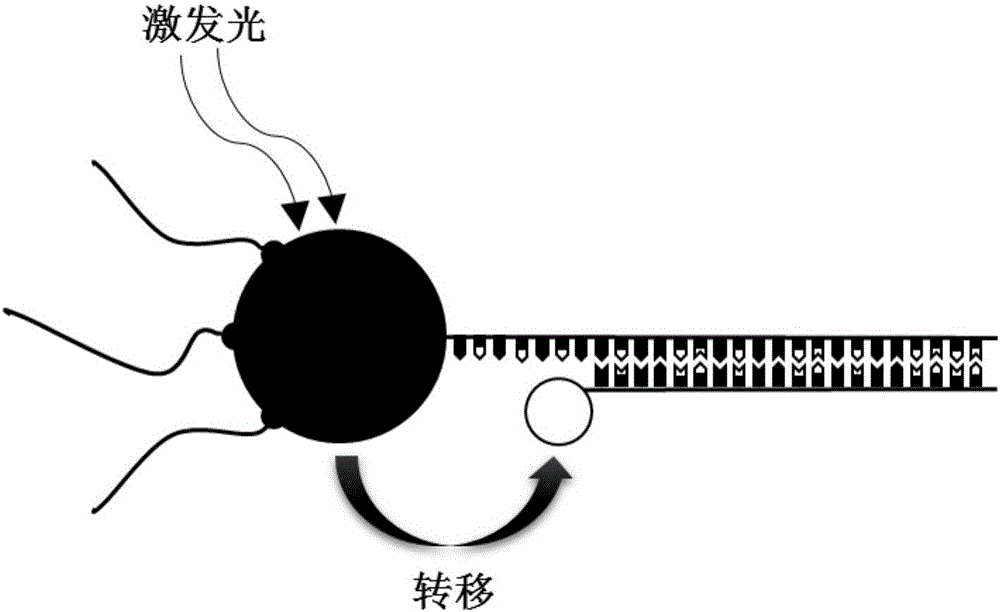
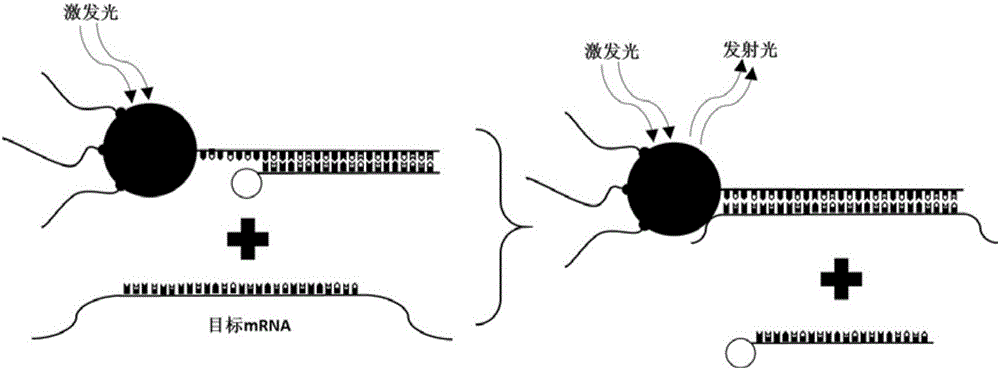
Nanoprobe for real-time parallel detection of content of various mRNA in living cells
A nanoprobe and living cell technology, applied in the field of nanoprobes, can solve the problems of cumbersome material synthesis process, expensive Raman spectroscopy detection equipment, complex pretreatment process, etc., and achieve the effect of strong anti-photobleaching performance
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0027] (1) Preparation of raw materials
[0028] Take 1pmol quantum dot QD605 (CdSe / ZnS quantum dot, emission wavelength is 605nm), connect 4-8 streptavidin on the periphery of each quantum dot;
[0029] The number of gold nanoparticles is 5-10 times the number of total binding sites of streptavidin; one streptavidin has 3 available binding sites, so the number of gold nanoparticles is 5-10 times that of streptavidin 15-30 times that of prime, the gold nanoparticles with a diameter of 2nm were used in the test;
[0030] Design and synthesize capture probes, reporter probes and penetrating peptides, and the synthesized probe sequences for K-ras gene and survivin gene are shown in Table 1. Wherein, the underlined part in the reporter probe represents the unpaired base at the end of the reporter probe, setting 2-3 unpaired bases at the end of the reporter probe can enhance the efficiency of competitive coupling; The underlined part is the targeting sequence region.
[0031] Ta...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com