Methods for multiplex detection of alleles associated with ophthalmic conditions
A genome, gene sequence technology, applied in the field of multiple detection of alleles related to ophthalmic conditions, which can solve problems such as vision loss, poor vision, and omission of potential symptoms of patients
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0225] Example 1: DNA extraction (DNA Extract All Reagents, ThermoFisher)
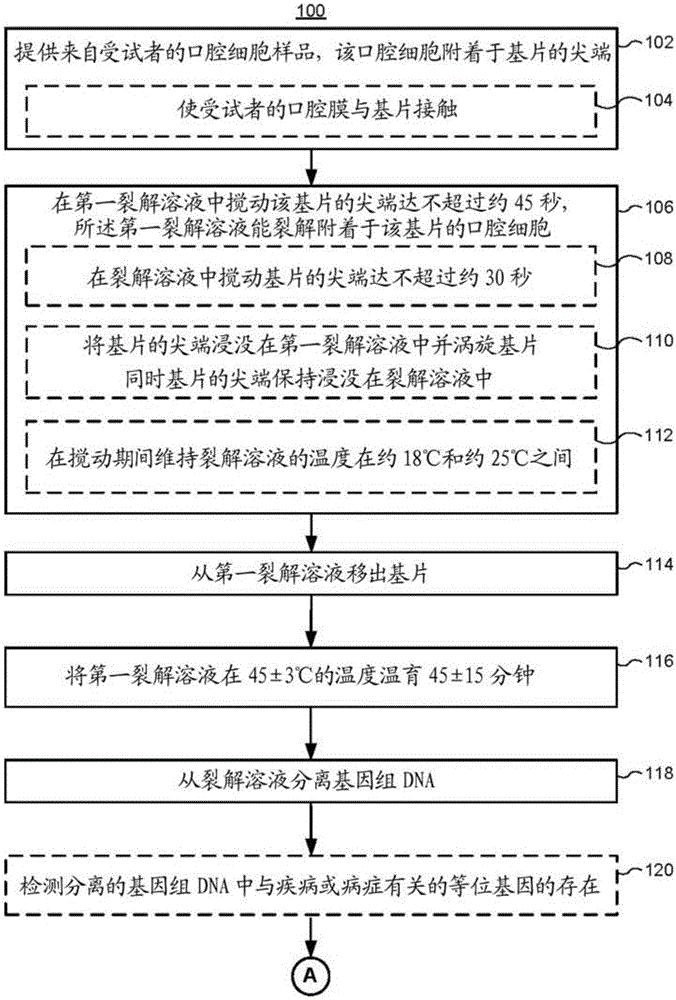
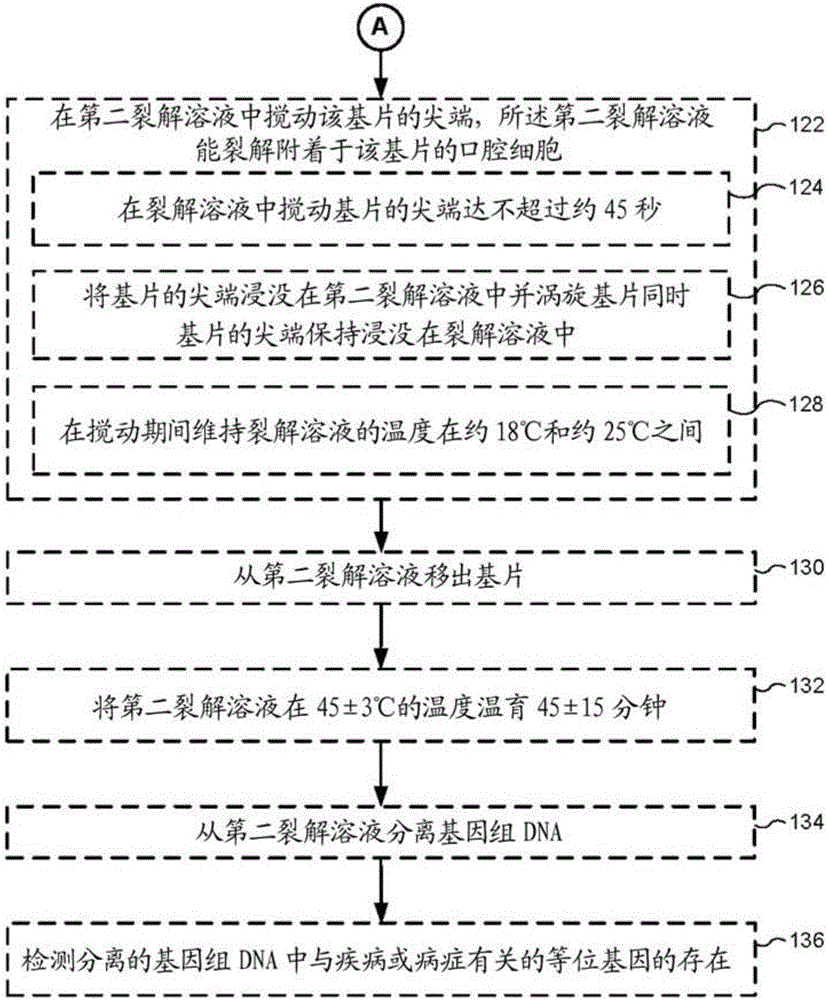
[0226] DNA was extracted from oral epithelium or hair roots or whole blood as described below and in accordance with the disclosure provided herein.
[0227] For DNA extraction from oral epithelium or hair roots, first pretreat samples in 300 µL of 1X PBS. Next, 30 μL of lysis solution was added to the tube and the mixture was vortexed. The mixture was then incubated at 95°C for 3 min. Then, 30 μL of DNA stabilization solution (from Life Technologies / Thermo Scientific, USA) was added and the mixture was vortexed. The mixture was then centrifuged at 13,000 RPM for 1 min.
[0228] For DNA extraction from whole blood, start with 3 µL of whole blood and first pretreat the sample in 300 µL of 1X PBS. Next, 30 μL of lysis solution was added to the tube and the mixture was vortexed. The mixture was then incubated at 95°C for 3 min. Then, 30 μL of DNA stabilization solution (from Life Technologies / Thermo S...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com