Gene mutation, deletion, insertion and recombinant fragment obtaining method based on overlapping extension PCR process
A technology of overlapping extension and acquisition method, which is applied in the fields of gene mutation, insertion and recombination fragment acquisition, and deletion, can solve the problems of difficulty in obtaining expected results, cumbersome operation, and difficulty in determining the cause, and achieves economical primer synthesis, simple operation and saving. Effects of time and reagents
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
specific Embodiment approach 1
[0024] Specific embodiment one: the method for obtaining gene mutation, deletion, insertion and recombination fragments based on the overlap extension PCR method of the present embodiment, it is carried out according to the following steps:
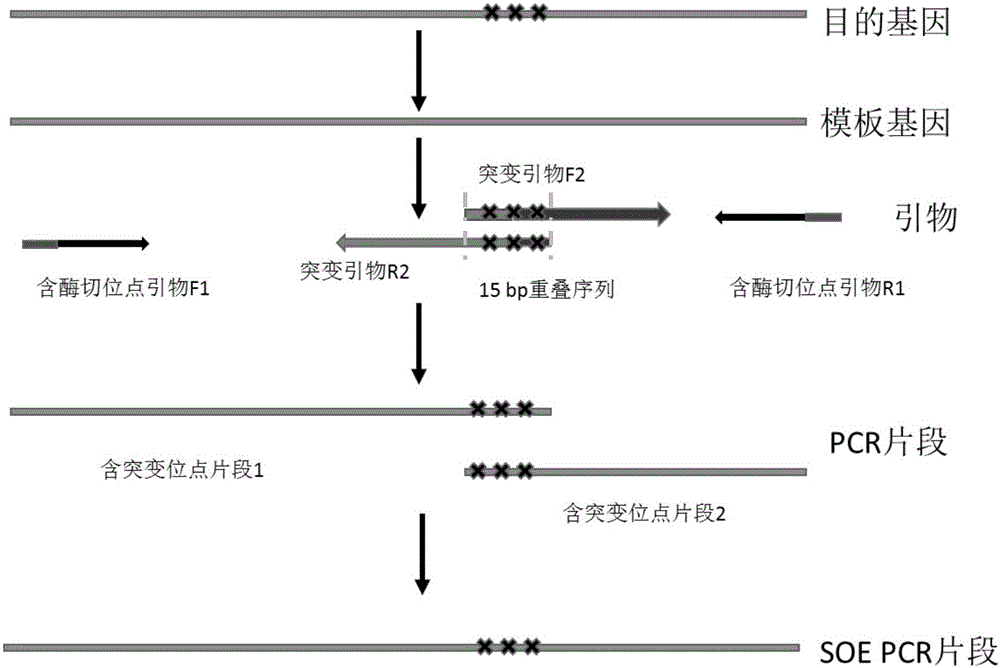
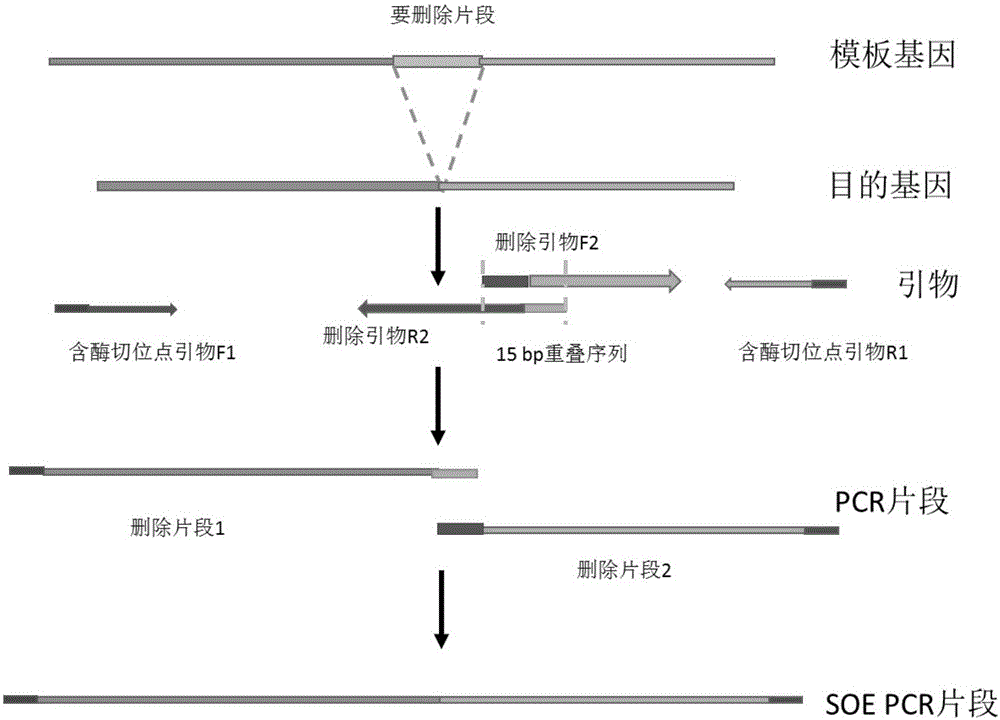
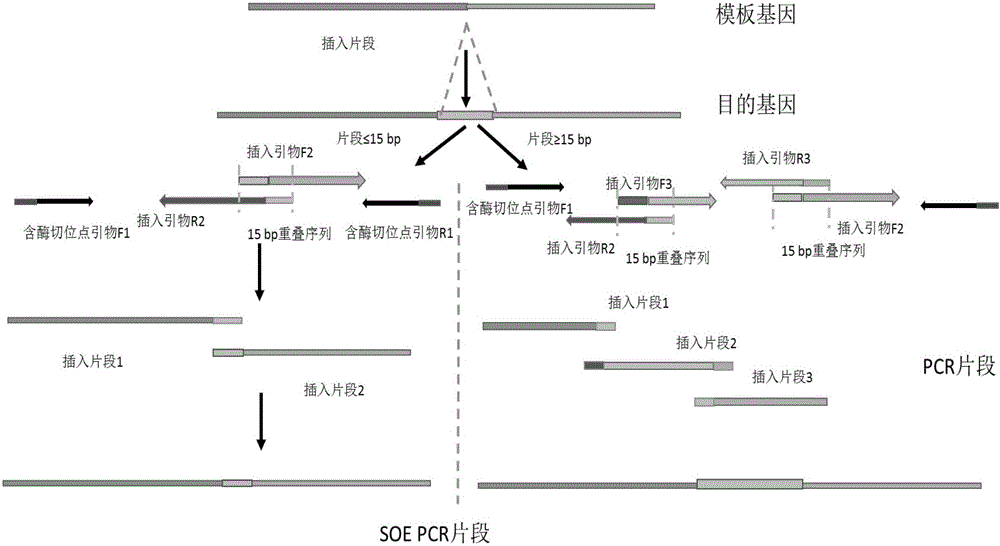
[0025] 1. Design two or three pairs of primers according to the sequence of the DNA fragments to be fused, and design an overlapping segment with a length of 15 bp at the 5' end of two adjacent primers designed corresponding to mutation, deletion, insertion and recombination sites, 3 The end of the primer is the specific binding sequence of the primer and the template, and then use the designed primers to carry out parallel PCR reactions with the gene fragment or plasmid DNA containing the desired fusion as the template, and obtain a fusion site containing 15 base overlaps. Two or three sets of PCR products;
[0026] 2. If the PCR product in step 1 is a single target band, take two or three sets of unpurified PCR products 1-2μl 2. If the ...
specific Embodiment approach 2
[0028] Specific embodiment 2: The difference between this embodiment and specific embodiment 1 is that in the process of obtaining gene mutation, the base site to be mutated is included in the 15 bp overlapping segment described in step 1 during primer design ( figure 1 ). Others are the same as in the first embodiment.
specific Embodiment approach 3
[0029]Specific embodiment 3: This embodiment differs from specific embodiment 1 in that in the process of obtaining gene deletion, the fragment to be deleted is deleted during primer design, and the 5' ends of the primers on both sides of the fusion site must be identified in step 1. The overlapping region of the 15bp fragment described above ( figure 2 ). Others are the same as in the first embodiment.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com