Method and kit for detecting one or more target sequence of multiple samples based on high-throughput sequencing
A target sequence, high-throughput technology, applied in the field of capturing and labeling one or more target sequences of multiple samples, methods or kits, can solve the problems of low throughput and high sequencing cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0070] 21 exons (EXON1, EXON2, EXON3, EXON4, EXON5, EXON6, EXON7, EXON8, EXON9, EXON10, EXON11, EXON12, EXON13, EXON14, EXON15, EXON10, EXON11, EXON12, EXON13, EXON14, EXON15, EXON16, EXON17, EXON18, EXON19, EXON20 and EXON21) sequencing, a total of 10 clinical samples:
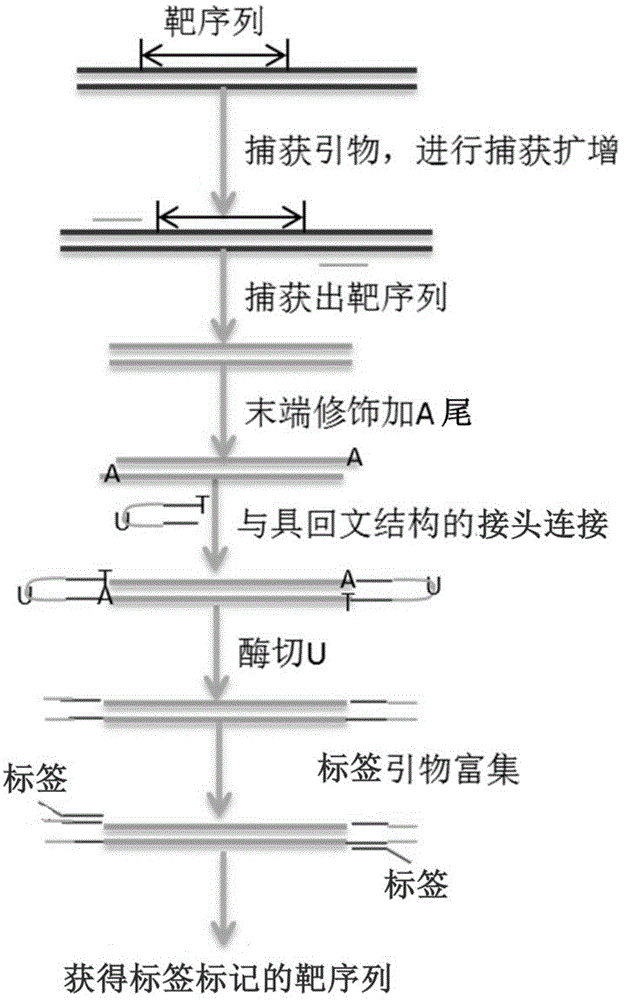
[0071] 1) Primer and linker design:
[0072] The corresponding capture primers were designed for the 21 exons of the ATP7B gene. Relevant parameters: Tm value 58.0℃-65.0℃, GC value 40.0%-60.0%, primer size 23±3bp, target fragment size 150-300bp, so The designed primers are as follows:
[0073]
[0074]
[0075] Design 2 adapters based on 10 samples, the adapters are single-stranded DNA, and the sequence contains a palindrome sequence of two regions (each region is 15bp in length), a U base and a 3'T end, which can form a stable hairpin structure , the designed linker sequence is as follows:
[0076] ADP1: 5'P-AGGACAGAAGCTCGA-U-TCGAGCTTCTGTCCT-s-T-3'
[0077] ADP2: 5'P-ACCTTGAGCGATCGA-U-TCGATCGCTCAAG...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com