Specific PCR primer, kit and detection method for detecting bovine HCD carrier
A kit and carrier technology, applied in the field of specific PCR primers and kits, can solve the problems of unclear carrier status of HCD haplotypes
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0033] Example 1 Primer Design
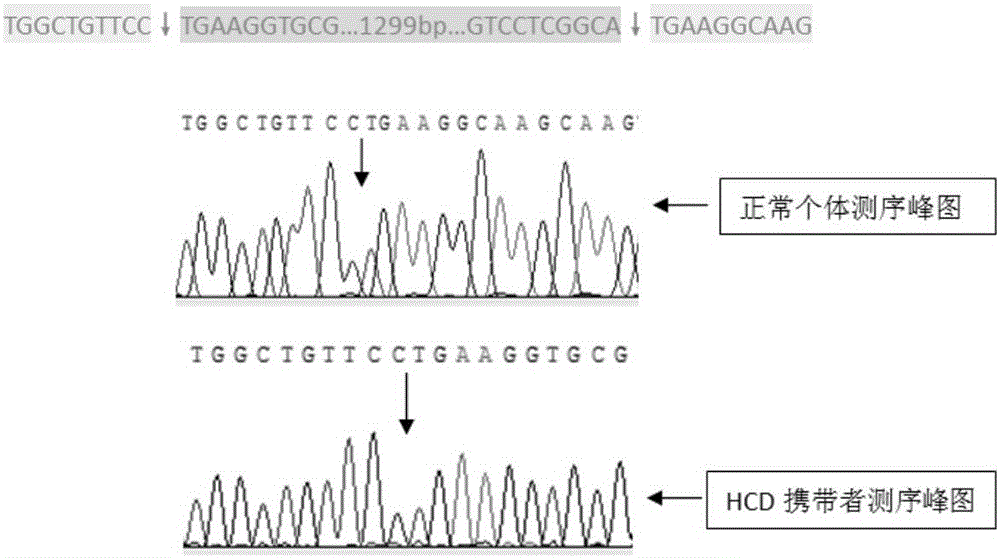
[0034] According to the bovine APOB gene sequence (ID: 494004) on GenBank, and referring to the primer sequence designed by Schütz et al. primer3-0.4.0 / ) to design specific primers for detecting HCD, and obtain primer pairs: upstream primer APOB-Primer-F: 5'-TGCAAAGCCCACCTAGCCTAT-3'; downstream primer APOB-Primer-R1: 5'-AGATGATGCCCCTCTTGATG-3 ', the primer pair spans the 1299bp insertion sequence of the APOB gene. Theoretically, normal individuals can amplify a 171bp fragment; HCD carrier individuals can amplify a 171 / 1470bp fragment, and HCD homozygotes can amplify A 1470bp length fragment was added. After the first round of primer screening and optimization, when the HCD carrier is amplified, the primer pair can only amplify a 171bp fragment, and the 1470bp length fragment cannot be amplified normally, which may be due to the 171bp short fragment Amplification inhibits the amplification of long fragments, and long fragments are inherently e...
Embodiment 2
[0046] Example 2 Optimization of PCR reaction program
[0047] Based on the designed specific PCR primers, the present invention further optimizes the reaction program of PCR.
[0048] Finally, the PCR reaction procedure and reaction system for identifying HCD carriers were determined.
[0049] Described reaction procedure is:
[0050] Pre-denaturation at 94°C for 5 minutes; 35 cycles of denaturation at 94°C for 30s, annealing at 62°C for 30s, and extension at 72°C for 30s; extension at 72°C for 7 minutes, and then storage at 4°C. Among them, the optimum annealing temperature is 62°C
[0051] The PCR reaction system: a total volume of 25 μL
[0052]
Embodiment 3
[0054] 1. Extraction of total bovine DNA
[0055] Holstein bulls from the Beijing Bull Breeding Station were selected as experimental materials to extract total DNA, which can be obtained from blood (for example, fresh or frozen), semen (for example, fresh or frozen), tissue samples (such as ear tissue, etc.) Or extraction, separation and purification from bovine hair samples containing hair follicles.
[0056] 2. PCR amplification and genotype determination
[0057] According to the specific primer pair screened in Example 1, and the PCR reaction conditions and reaction system optimized in Example 2, multiplex PCR amplification was performed.
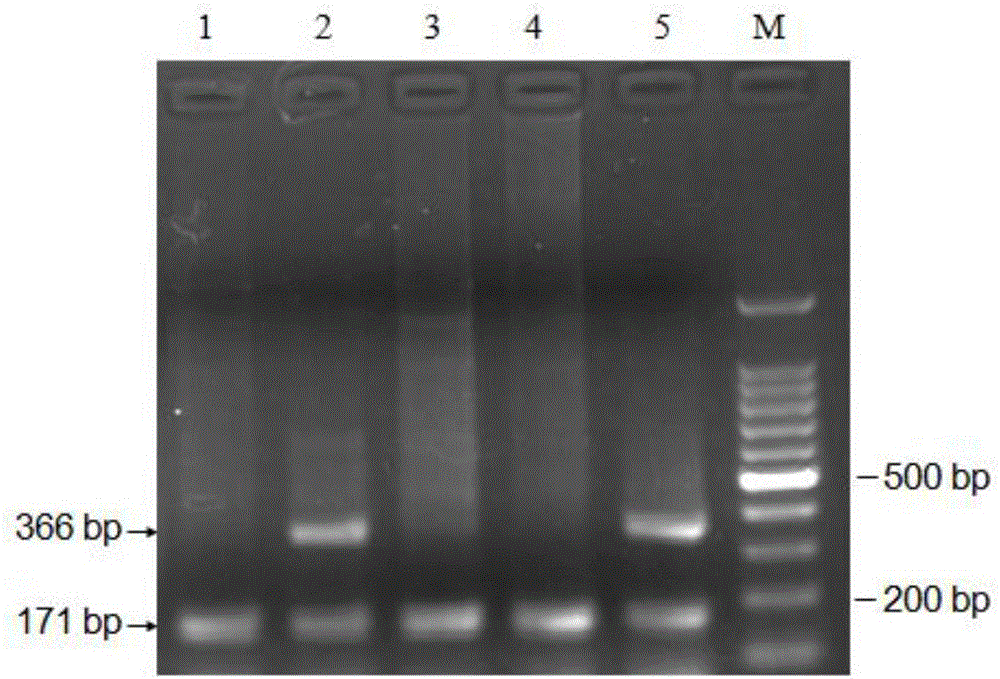
[0058] Take 4 μL of PCR products and detect them by 2% agarose gel electrophoresis. The results of PCR amplification are shown in the attached figure 1 shown.
[0059] Genotype determination:
[0060] Healthy dairy cows: the genotype is AA, and only 171bp electrophoresis band appears;
[0061] HCD recessive carrier: the genotype i...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com