Parallel rapid matching method and system for stored DNA sequence
A technology of DNA sequence and matching method, which is applied in the field of parallel fast matching method of DNA sequence and its system, which can solve the problem of low efficiency of DNA sequence matching and achieve the effect of improving efficiency and speeding up operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0058] In order to make the object, technical solution and advantages of the present invention clearer, the present invention will be further described in detail below in conjunction with the accompanying drawings and embodiments. It should be understood that the specific embodiments described here are only used to explain the present invention, not to limit the present invention.
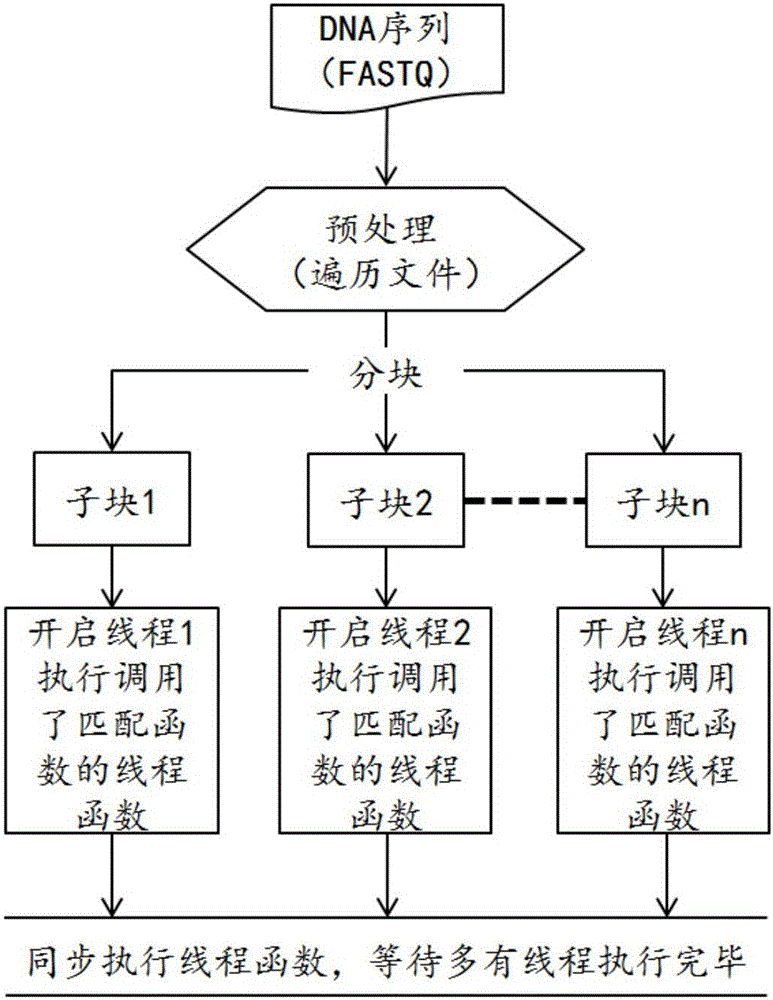
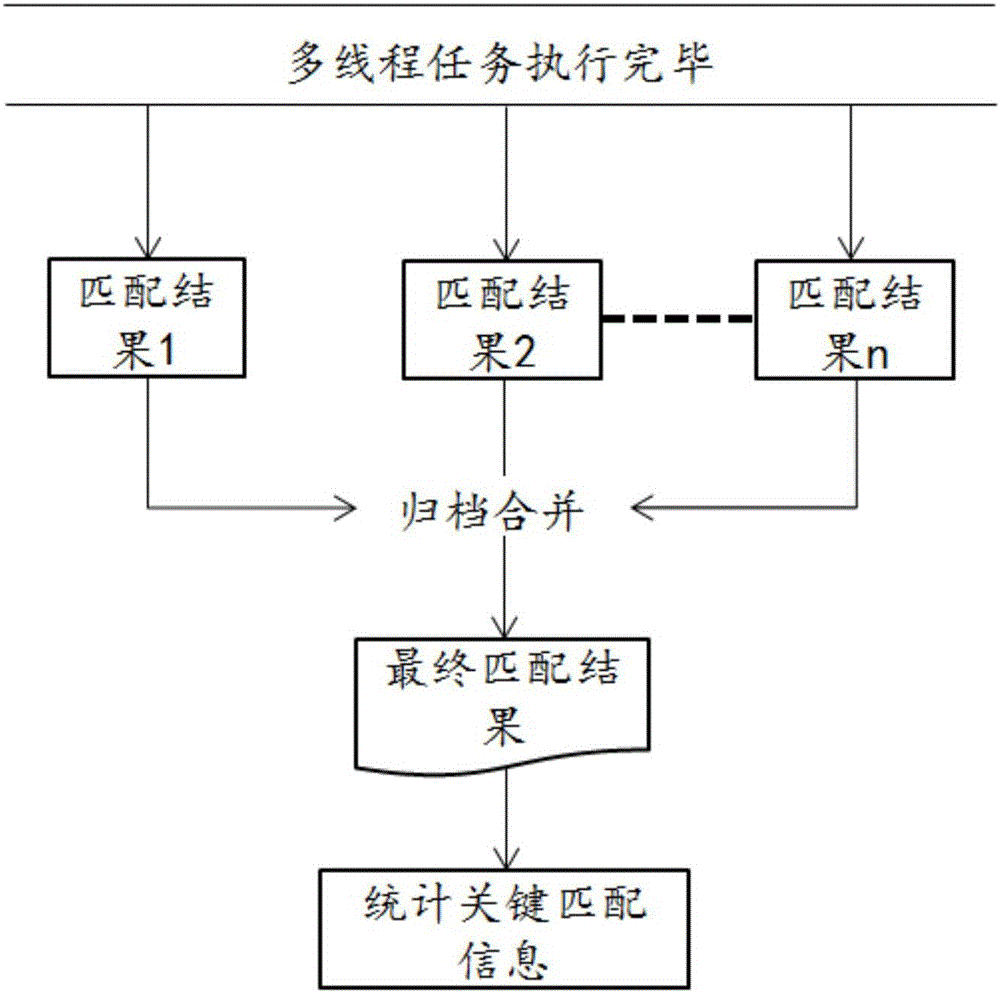
[0059] The specific embodiment of the present invention provides a parallel fast matching method for stored DNA sequences, which is applied to the compressed storage of DNA sequences, wherein the method mainly includes the following steps:
[0060] S11. Hash index construction step: construct a hash index based on the reference genome in FASTA format based on the prefix, find out all the kmers with the specified prefix and use them as key values to build a hash index table, and each entry stores the position where the kmer appears ;
[0061] S12, file block step: input the DNA sequence file in F...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com