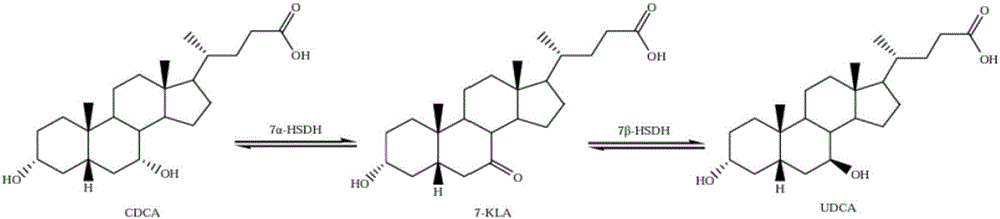
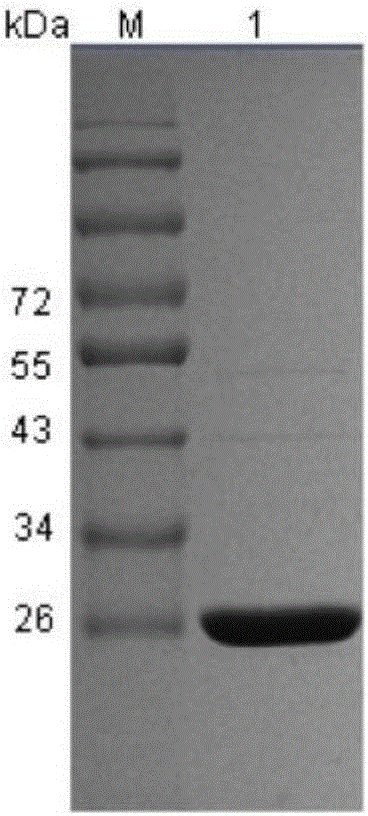
Clostridium sardiniense 7alpha-hydroxysteroid dehydrogenase mutant T145S
A technology of hydroxysteroids and dehydrogenases, applied in bacteria, genetic engineering, oxidoreductases, etc., can solve problems such as low stereoselectivity, difficult recovery, limited types and numbers of catalysts, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0036] Example 1. Preparation of Clostridium sardinia 7α-hydroxysteroid dehydrogenase mutant
[0037] 1. Mutant Gene Synthesis
[0038] Original sequence: codon-optimized wild-type CA 7α-HSDH gene sequence (see patent publication CN102827848A), the nucleotide sequence of which is shown in SEQ ID NO:4.
[0039] By comparing the similarities and differences between wild-type CA 7α-HSDH and homologous enzyme proteins from the primary structure to the high-order structure, the site that affects the enzymatic properties is the 145th amino acid of wild-type CA 7α-HSDH , the amino acid is threonine, and the corresponding nucleotide sequence is codons 433-435.
[0040]The codons 433-435 of the wild-type CA 7α-HSDH gene sequence were changed from ACT to AGT, and the original threonine was replaced with serine to obtain a CA 7α-HSDH mutant, named CA 7α-HSDH T145S mutant , its nucleotide sequence is shown in SEQ ID NO: 3, and its amino acid sequence is shown in SEQ ID NO: 2.
[0041] ...
Embodiment 2
[0062] Example 2. Determination of T145S Mutant Enzyme Kinetic Parameters
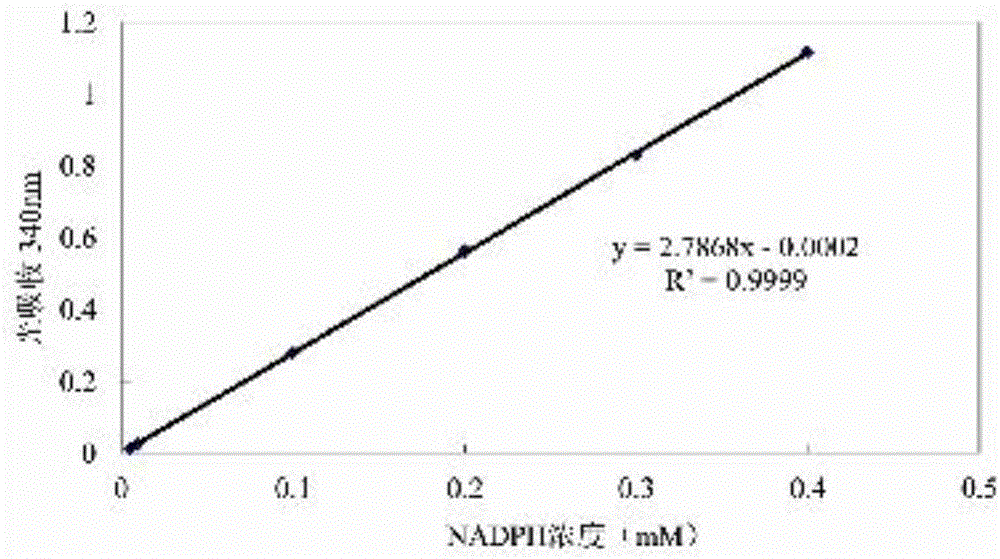
[0063] 1. Preparation of NADPH standard curve
[0064] Using reaction buffer (50 mM Tris-HCl, 200 mM NaCl, pH 8.0), 0.1, 0.2, 0.3 and 0.4 mM NADPH solutions were prepared respectively. After zeroing with the above blank solvent, add NADPH solutions of various concentrations into 2mL cuvettes respectively, and measure the light absorption value OD at 340nm 340 . With the concentration of NADPH solution as the abscissa and the corresponding light absorption value as the ordinate, a standard curve was drawn.
[0065] The results are shown in Figure 3, the obtained standard curve equation is y=2.7868x-0.0002, R 2 = 0.9999.
[0066] 2. Enzyme activity assay
[0067] Add 958μL of reaction buffer (50mM Tris-HCl, 200mM NaCl, pH 8.0), 20μL of 50mM NADP to a 2mL cuvette + . 2 μL of the CA 7α-HSDH T145S mutant (2.46 mg / mL) prepared in Example 1, after zero adjustment, 20 μL of 50 mM substrate TCDCA was adde...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com