Primers for rapidly identifying bemisia tabaci sodium ion channel gene mutation T929V and application thereof
A sodium ion channel and Bemisia tabaci technology, applied in the field of molecular biology, can solve problems such as the design of specific primers that cannot be fully applied, and achieve the effect of improving the accuracy rate
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
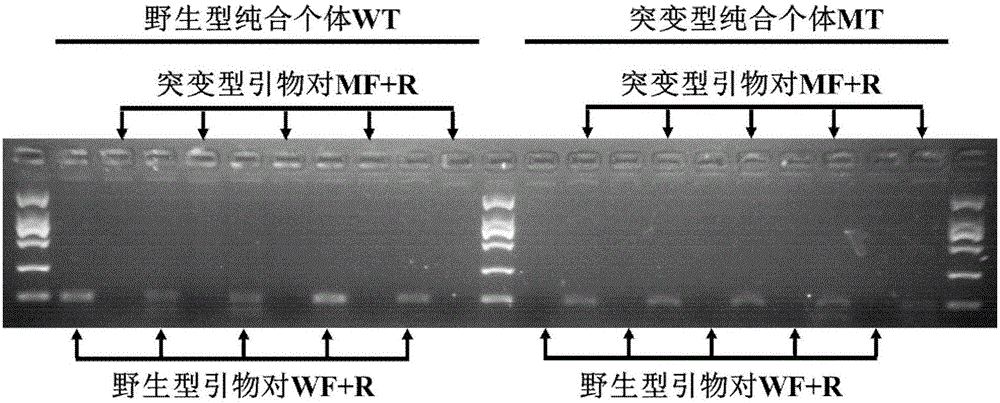
[0043] Sequencing verification of wild-type and mutant templates of sodium ion channel gene T929V in Bemis tabaci
[0044] Design specific primers according to the Bemisia tabaci sodium ion channel gene (Genbank: DQ205209.1) published by NCBI, perform PCR amplification on the 929-position mutation, and directly perform PCR sequencing after recovering the specific target fragment.
[0045] Primers for PCR amplification:
[0046] Upstream primer: GGCCAACTTTGAATCTGTT;
[0047] Downstream primer: AAACTTTCCGCACCTCTG.
[0048] PCR amplification system: Genomic DNA solution 2μl, 10M upstream primer 1μl, 10M downstream primer 1μl, 5U / μl rTaq enzyme 0.25μl, 10×Taq Buffer (buffer) 2.5μl, 10mM dNTP 2μl, ddH 2 0 to 25 μl;
[0049] PCR amplification conditions: pre-denaturation at 94°C for 3 minutes; denaturation at 94°C for 30 seconds, annealing at 50°C for 30 seconds, extension at 72°C for 30 seconds, and 35 cycles; extension at 72°C for 7 minutes.
[0050] According to the above, th...
Embodiment 2
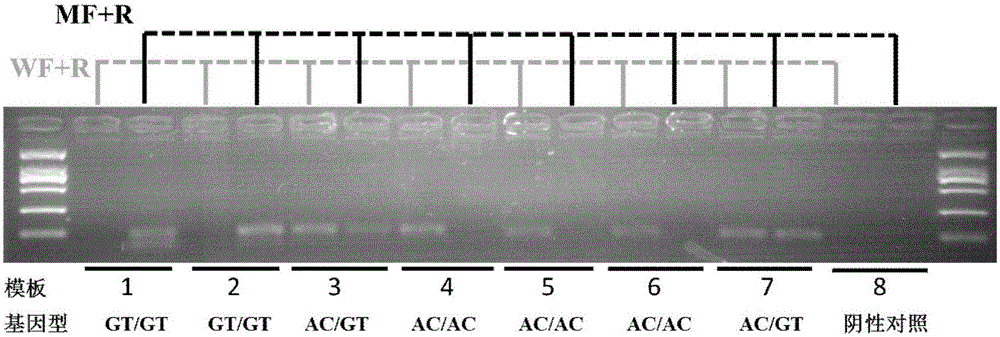
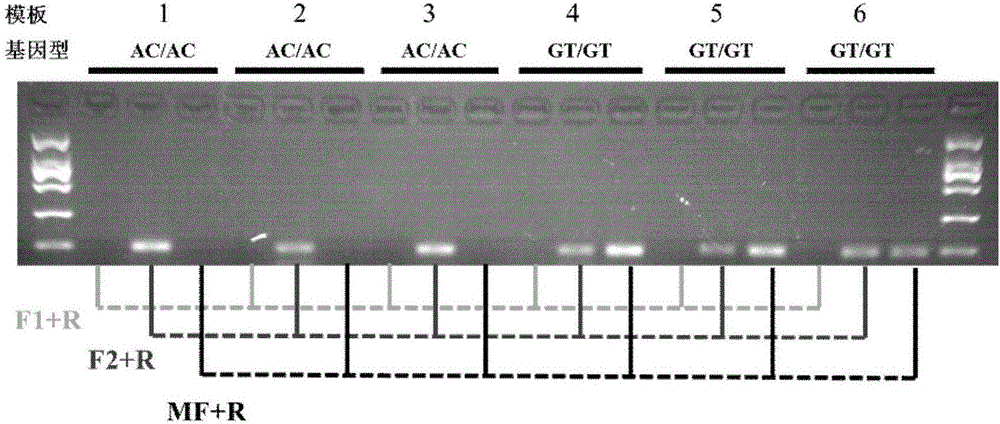
[0065] A primer for quickly identifying the mutation site T929V of the sodium ion channel gene as wild-type Bemisia tabaci, the primers are a pair, the nucleotide sequence of the upstream primer is shown in SEQ ID NO.1, and the nucleotide sequence of the downstream primer is shown in SEQ ID NO.1 Shown in ID NO.2.
[0066] A primer for quickly identifying the mutation site T929V of the sodium ion channel gene as a mutant whitefly, the primers are a pair, the nucleotide sequence of the upstream primer is shown in SEQ ID NO.3, and the nucleotide sequence of the downstream primer is shown in SEQ ID NO.3 Shown in ID NO.2.
[0067] The method for identifying the T929V genotype of the Bemisia tabaci sodium ion channel gene mutation site using the above primers, the steps are as follows:
[0068] (1) extracting the genomic DNA of the sample to be identified to obtain a genomic DNA solution;
[0069] (2) Using the genomic DNA obtained in step (1) as a template, use the primers of the...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com