Method and special primers for rapidly detecting cowpea thrips sodium ion channel I904S mutation
A sodium ion channel and ion channel technology, applied in the field of rapid detection of the I904S mutation of the cowpea thrips sodium ion channel, can solve the problems of time-consuming, unsuitable for detection, and heavy workload, and achieve the effect of delaying the development of drug resistance
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
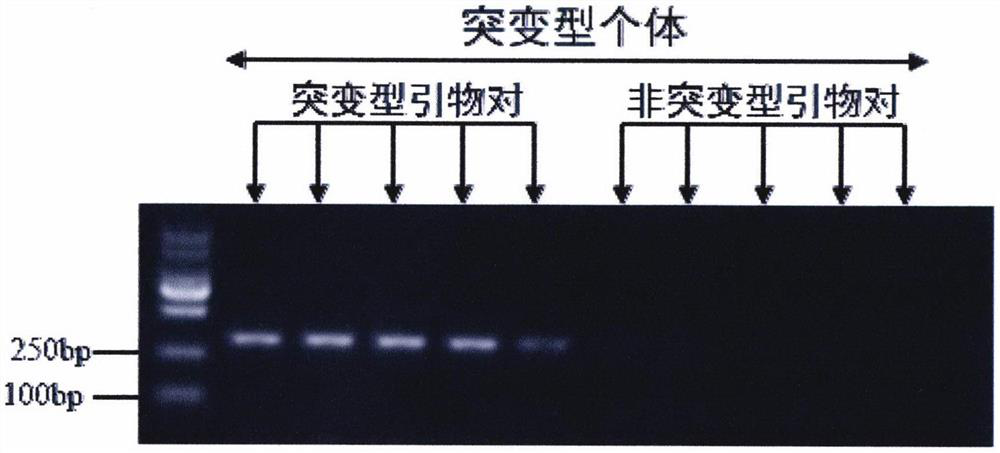
[0030] A method for rapid detection of the I904S mutation of the cowpea thrips sodium ion channel and special primers, the primers are a pair: the nucleotide sequence of the upstream primer is shown in SEQ ID NO.2, and the nucleotide sequence of the downstream primer is shown in The nucleotide sequence shown in SEQ ID NO.3; the nucleotide sequence shown in SEQ ID NO.1 is the upstream nucleotide sequence of the thrips cowpea without mutation.
[0031] Utilize above-mentioned primer to identify the method that mutation site I904S of cowpea thrips sodium ion channel gene mutation site is as follows:
[0032] (1) Primer design
[0033] The nucleotide sequences of the primers are shown in Table 1, and allele specific-PCR primers were designed according to the gene sequence of the mutant cowpea thrips sodium ion channel domain II:
[0034] The last base at the 3' end of the dedicated unmutated primer pair is consistent with the base sequence before the mutation (SEQ ID NO.1);
[0...
Embodiment 2
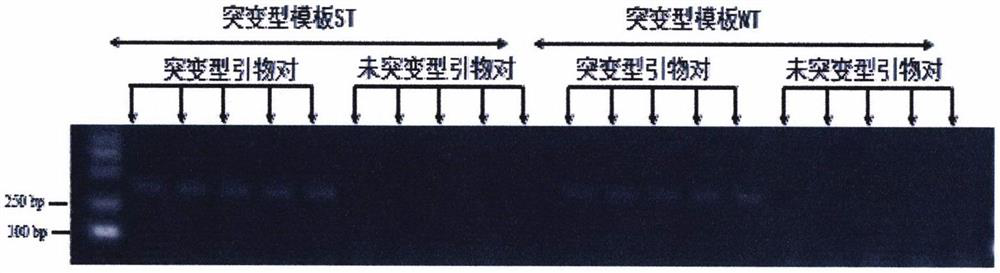
[0048] Sequencing Verification of I904S Mutant Template of Sodium Channel Gene of Vigna thrips
[0049] Specific primers were designed according to the sodium ion channel gene of Vigna grandis thrips (GenBank accession number: MT506055), and the 904-position mutation was amplified by PCR, and the specific target fragment was recovered and sequenced directly.
[0050] PCR amplification primers:
[0051] Upstream primer: SEQ ID NO.2
[0052] Downstream primer: SEQ ID NO.3
[0053] PCR amplification system: pre-denaturation at 94°C for 3 min; denaturation at 94°C for 15 s, annealing at 52°C for 15 s, extension at 72°C for 15 s, and 30 cycles; extension at 72°C for 8 min.
[0054] According to the above, the PCR amplification of the two field populations was completed, and after the agarose gel electrophoresis test analysis, any samples with bands at 270bp were randomly selected for sequencing. The sequencing results are as follows:
[0055] Individual DNA mutation template WT ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com