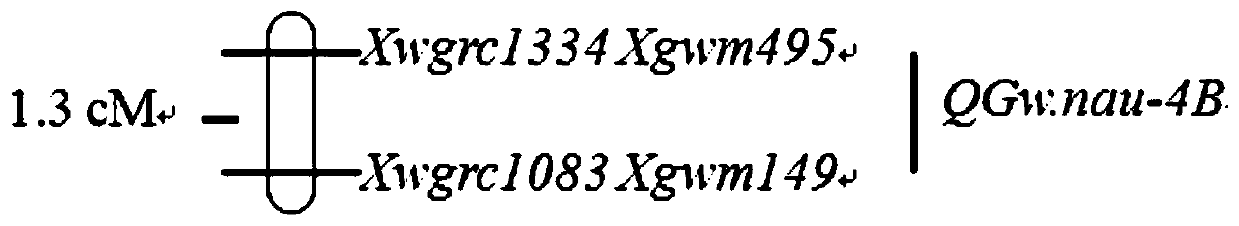Molecular Marker Primer and Its Application of qgw.nau-4b Main Effect Gene Locus of Wheat Grain Weight
A qgw.nau-4b, molecular marker technology, applied in the field of crop breeding, can solve problems such as thousand-grain weight difference, and achieve the effects of high cost, saving breeding cost, and stable amplification
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0027] The molecular markers of wheat grain weight main gene QGw.nau‐4B were obtained by the following methods:
[0028] According to the boundary markers Xgwm495 and Xgwm149 of QGw.nau‐4B (Jia et al 2013), the published wheat genome sequence is located in the QGw.nau‐4B segment (http: / / wheat.pw.usda.gov / GG2 / index. shtml) and Wangshuibai BAC clone sequence to screen the polymorphisms of Nanda 2419, Wenmai 6 and Wangshuibai, and finally get two polymorphic markers between the parents, namely WGRC1334 and WGRC1083 Two molecular markers.
[0029] The PCR reaction system is 12.5μl, including 1.25μl of 10×buffer, 25mM MgCl 2 0.75μl, 2.5mMdNTPs 1μl, upstream and downstream primers 0.2μM each, Taq enzyme (5u / μl) 0.1μl, template DNA 10ng, add water to 12.5μl;
[0030] The PCR amplification program was pre-denaturation at 94°C for 3 min, denaturation at 94°C for 30 sec, annealing at 60°C for 1 min, extension at 72°C for 1 min, 35 cycles, and finally 8 min at 72°C; The products were...
Embodiment 2
[0032] Example 2 Molecular marker-assisted transfection of QGw.nau-4B segment increases wheat grain weight
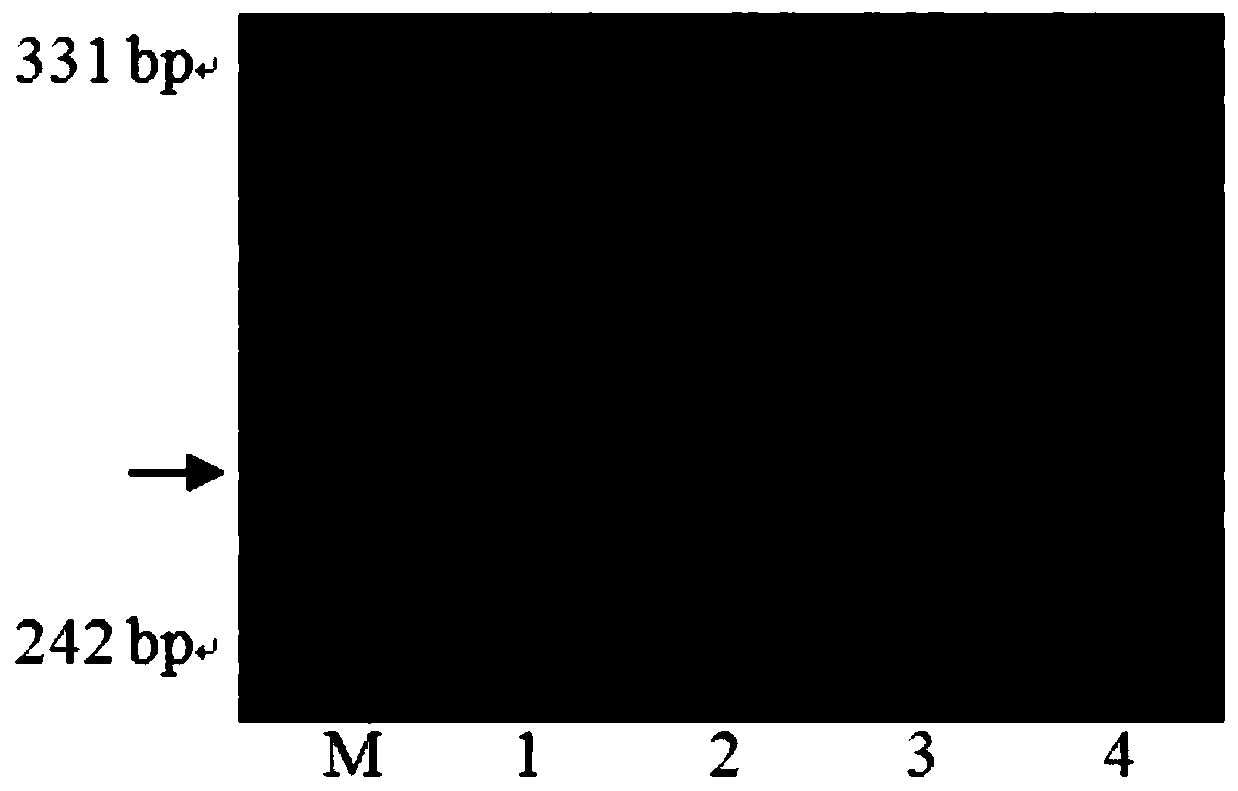
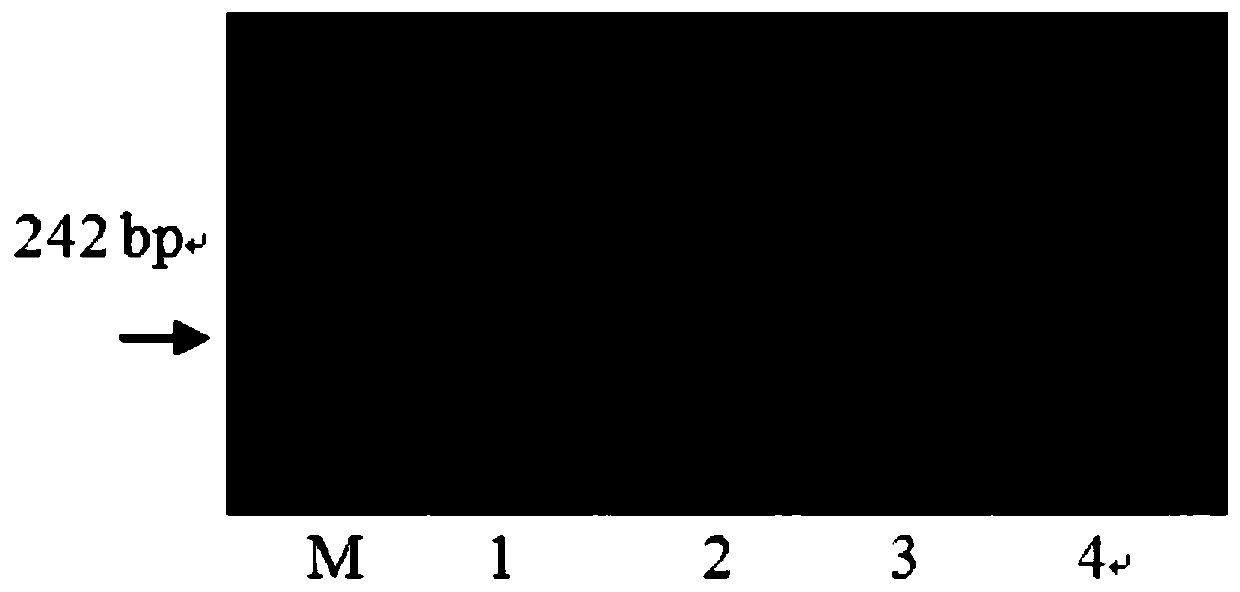
[0033] Using the homozygous near-isogenic genomic DNA of Wenmai No. 6 background as a template, two pairs of molecular marker primers were used to amplify by PCR and detect the amplified products. The amplified fragment of 220bp, and the primer WGRC1083-F and WGRC1083-R of molecular marker WGRC1083 were used to amplify the amplified fragment of 265bp, indicating that the QGw.nau-4B segment was detected in the DNA of the near-isogenic line. Molecular marker primer amplification band patterns see figure 2 and image 3 . Based on the genotypes, identify the grain weight phenotypes of near-isogenic lines and their recipient parents. The results showed that with Wenmai 6 as the control, the 100-kernel weight of the near-isogenic line was significantly increased (Table 1).
[0034] Table 1 The genotype and phenotype of the QGw.nau‐4B near-isogenic line and Wenmai 6 in th...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com