Method for predicting protein bound with ribonucleic acid
A prediction method and ribonucleic acid technology, applied in proteomics, genomics, instruments, etc., can solve the problem of low accuracy of RBP, and achieve the effect of reducing the amount of calculation, accurate prediction model, and comprehensive and accurate prediction
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
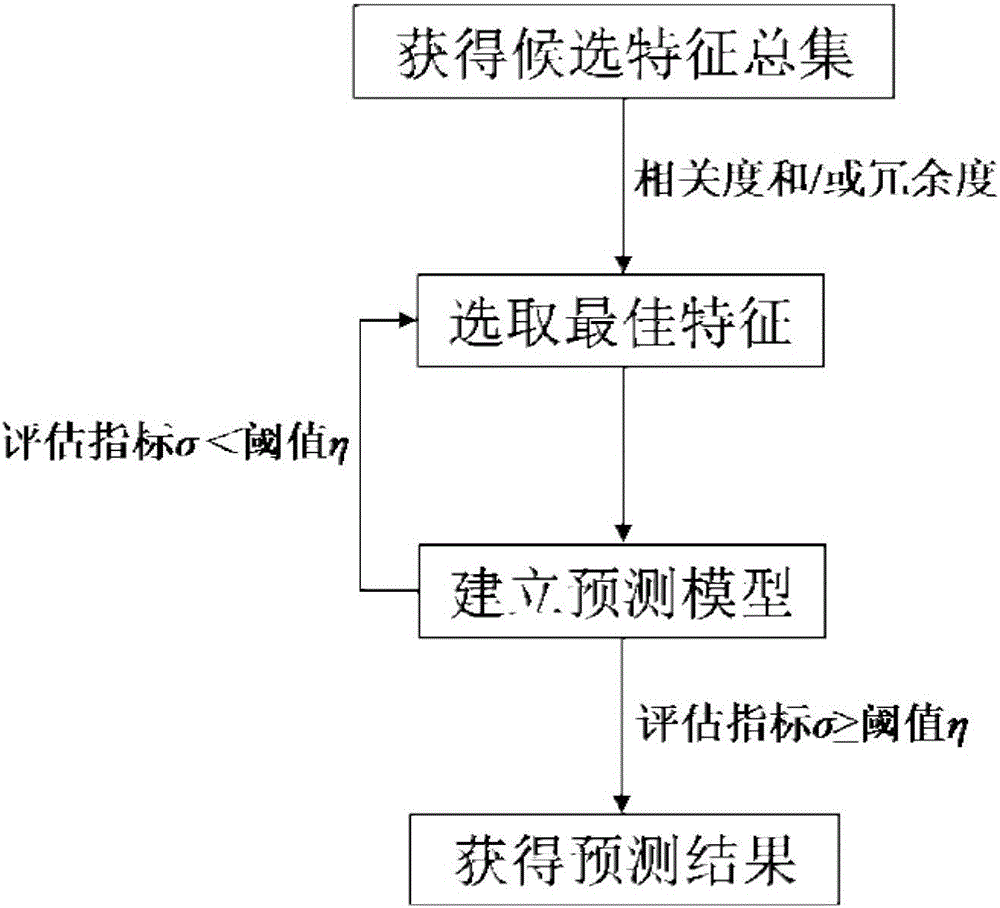
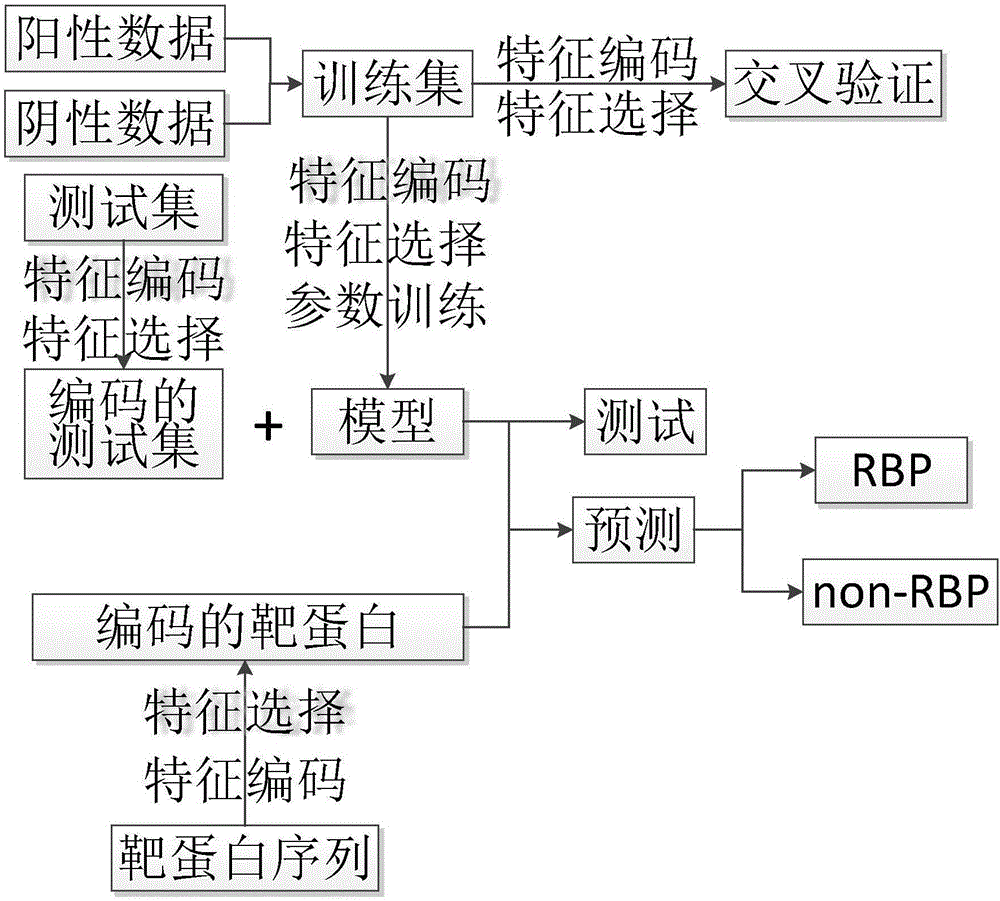
[0118] The prediction method of embodiment 1 includes the selection of feature vectors, the construction of prediction models and the prediction of the target protein to be predicted;
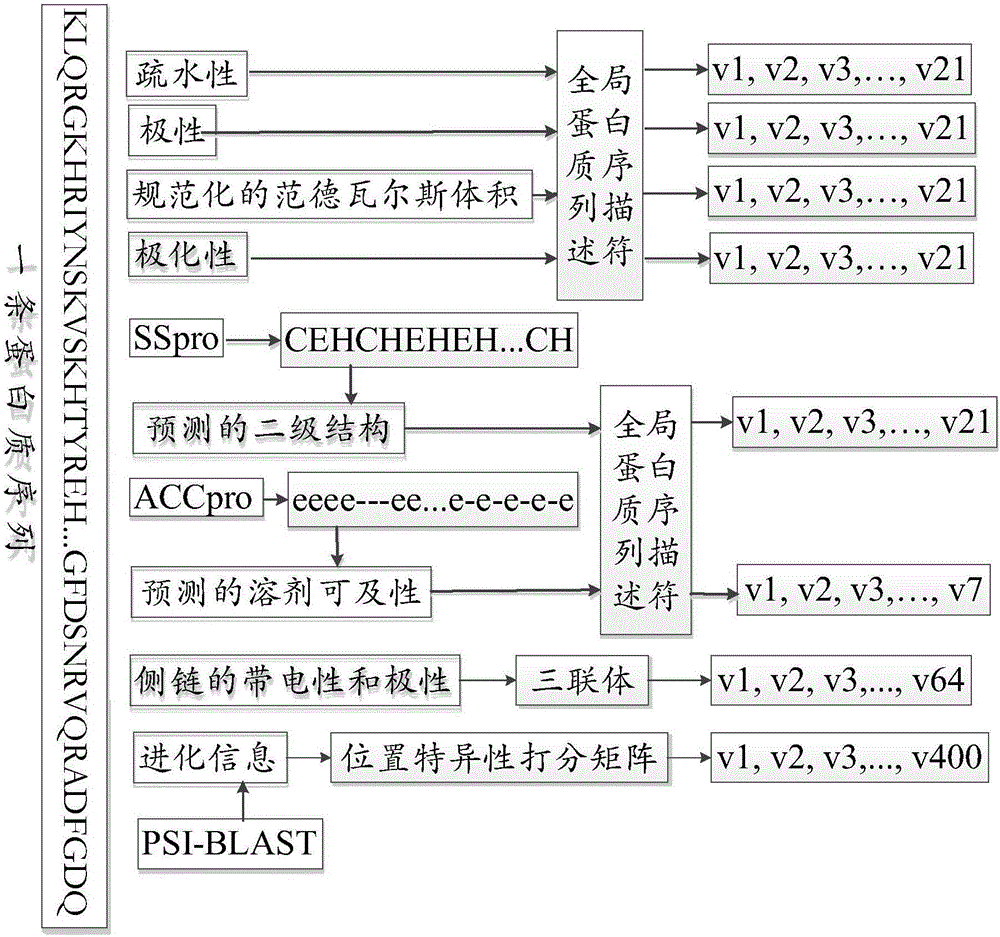
[0119] S1. figure 2 It is a flow chart of selecting eigenvectors in Embodiment 1 of the invention, including the first eigenvector to the eighth eigenvector; respectively corresponding to the hydrophobicity of the amino acids contained in the protein, the polarity of the amino acids contained in the protein, and the normalized range of the amino acids contained in the protein De Waals volume, the polarizability of amino acids contained in proteins, the secondary structure of proteins, the solvent accessibility of proteins; the charge and polarity of the side chains of amino acids contained in proteins; and the evolution information of proteins;
[0120] S11. Wherein, the first eigenvector to the sixth eigenvector are obtained by using the encoding method of the global protein sequence descript...
Embodiment 2
[0187] According to the above steps of S4, the human protein dataset was tested as the target protein, and the human protein dataset included 967 RBPs and 579 non-redundant non-RBPs. The prediction results are as follows: 84% of 967 RBPs are correctly predicted, 97% of 597 non-RBPs are correctly predicted, and the Matthew correlation coefficient is 0.788.
Embodiment 3
[0189] Repeat Example 2 with the same steps described above, the difference is that the target protein is the protein data set of yeast, and the corresponding Matthew correlation coefficient is 0.729.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com