Tumor key gene identification method based on prior information and parallel binary particle swarm optimization
A technology of particle swarm optimization and prior information, which is applied in the field of tumor key gene identification based on prior information and parallel binary particle swarm optimization, can solve the problem of few samples of gene expression profile data, no corresponding reports on tumor gene identification, gene expression Solve the problems of high noise and high variation data analysis of spectral data, so as to improve the effect of tumor identification
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
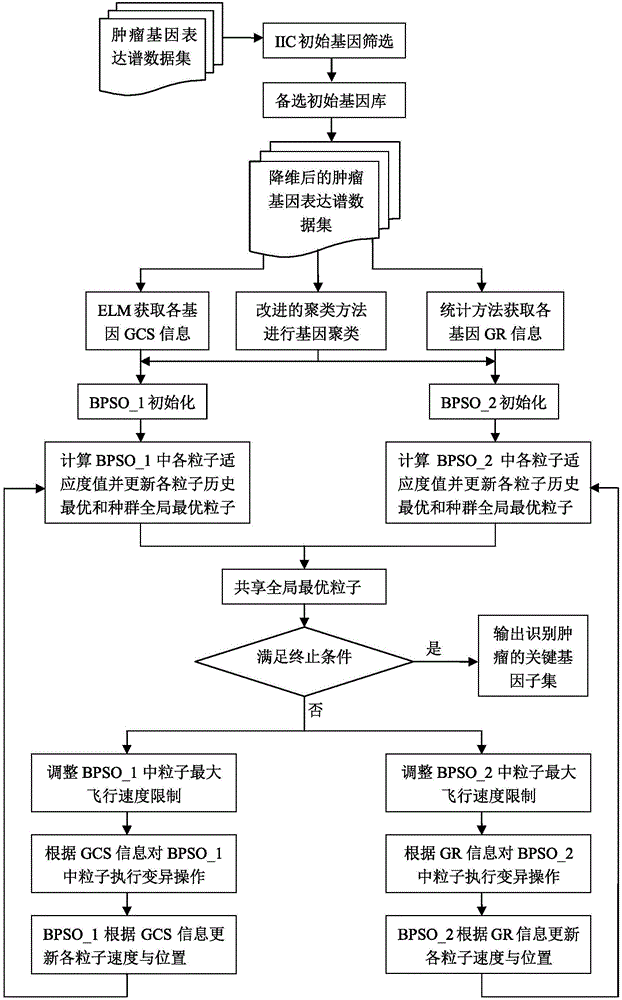
[0052] A tumor key gene identification method based on prior information and parallel binary particle swarm optimization, including K-means gene clustering based on particle swarm optimization (PSO), and using prior information and parallel binary particle swarm optimization (Binary Particle Swarm Optimization, BPSO) carries out the step of key gene identification, and the present invention specifically comprises the following steps:
[0053] Step 1. Preprocessing of tumor gene expression profile data, including normalization and preliminary dimensionality reduction of tumor gene expression profile data sets, and simultaneously dividing tumor gene expression profile data sets into training sets and test sets;
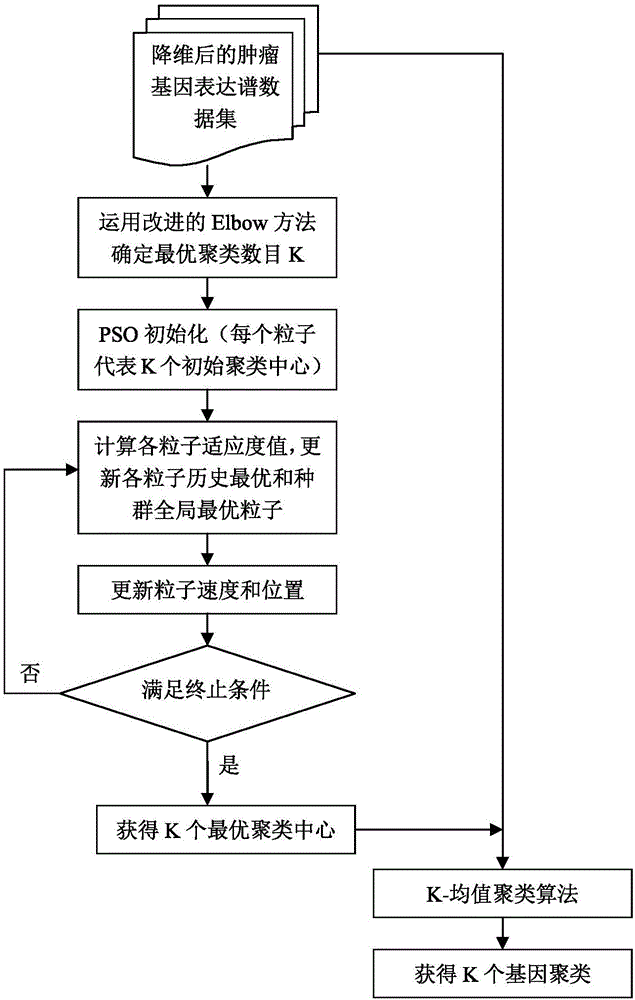
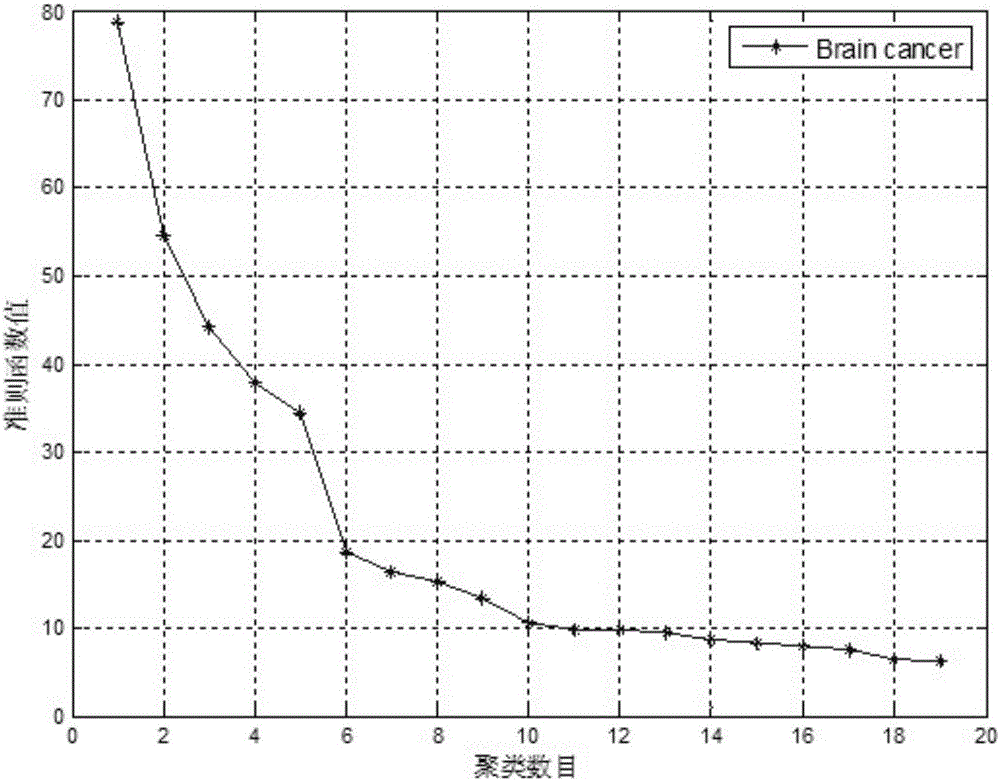
[0054] Step 2: On the training set, use the improved Elbow method to determine the optimal gene clustering number K through the self-defined criterion function;
[0055] Step 3: Use the PSO algorithm to select K optimal cluster centers, and use the K-means method on the...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com